Antibacterial, Antihemolytic, Cytotoxic, Anticancer, and Antileishmanial Effects of Ajuga bracteosa Transgenic Plants
Abstract
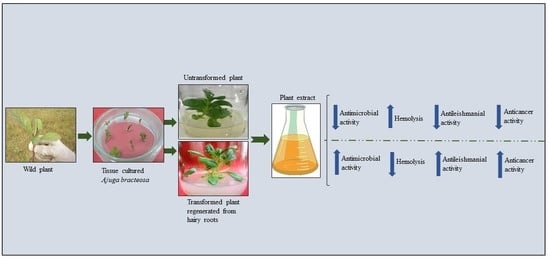
:1. Introduction
2. Results
2.1. Molecular Analyses of Ajuga bracteosa Plants
2.2. FTIR Coupled with PCA
2.3. Antibacterial Activity
2.4. The Antihemolytic Activity of Plant Extracts
2.5. Brine Shrimp Lethality Activity
2.6. Plant Extracts Inhibited Cancer Cell Growth in a Concentration-Dependent Manner
2.7. Antileishmanial Activity
3. Discussion
4. Material and methods
4.1. Plant Source
4.2. Confirmation of Genetic Integration by PCR and RT-PCR
4.3. Crude Extract Preparation
4.4. Identification of Functional Groups by Fourier Transform Infra-Red (FTIR) Spectroscopy
4.5. Antibacterial Activity
4.6. Anti-Hemolytic Assay
4.7. Brine Shrimp Cytotoxicity Assay
4.8. Antiproliferative Activity
4.8.1. 3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyl-2H-Tetrazolium Bromide (MTT) Assay
4.8.2. Sample Preparation for MTT Assay
4.8.3. Maintenance of Cell Cultures
4.8.4. Assay Procedure
4.9. Anti-Promastigote Assay
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Bisi-Johnson, M.A.; Obi, C.L.; Samuel, B.B.; Eloff, J.N.; Okoh, A.I. Antibacterial activity of crude extracts of some South African medicinal plants against multidrug resistant etiological agents of diarrhoea. BMC Complement. Altern. Med. 2017, 17, 321. [Google Scholar] [CrossRef] [PubMed]
- Adamczak, A.; Ożarowski, M.; Karpiński, T.M. Antibacterial activity of some flavonoids and organic acids widely distributed in plants. J. Clin. Med. 2020, 9, 109. [Google Scholar] [CrossRef] [Green Version]
- Granato, D.; Mocan, A.; Câmara, J.S. Is a higher ingestion of phenolic compounds the best dietary strategy? A scientific opinion on the deleterious effects of polyphenols in vivo. Trends Food Sci. Technol. 2020, 98, 162–166. [Google Scholar] [CrossRef]
- García-Becerra, L.; Mitjans, M.; Rivas-Morales, C.; Verde-Star, J.; Oranday-Cárdenas, A.; María, P.V. Antioxidant comparative effects of two grape pomace Mexican extracts from vineyards on erythrocytes. Food Chem. 2016, 194, 1081–1088. [Google Scholar] [CrossRef]
- Balderrama-Carmona, A.P.; Silva-Beltrán, N.P.; Gálvez-Ruiz, J.C.; Ruíz-Cruz, S.; Chaidez-Quiroz, C.; Morán-Palacio, E.F. Antiviral, antioxidant, and antihemolytic effect of Annona muricata L. leaves extracts. Plants 2020, 9, 1650. [Google Scholar] [CrossRef] [PubMed]
- da Silva, A.C.; do Nascimento, R.M.; do Nascimento Rodrigues, D.C.; Ferreira, P.M.; Pessoa, C.; Lima, D.J.; de Moraes Filho, M.O.; de Almeida, R.M.; Ferreira, S.R.; Fujiwara, R.T.; et al. In vitro activity evaluation of seven Brazilian Asteraceae against cancer cells and Leishmania amazonensis. S. Afr. J. Bot. 2019, 1, 267–273. [Google Scholar] [CrossRef]
- World Health Organization. Cancer-WHO. Available online: http://www.who.int/cancer/en (accessed on 14 August 2021).
- The Cancer Atlas. Southern, Eastern, and South-Eastern Asia. Available online: https://canceratlas.cancer.org/the-burden/south-east-se-asia (accessed on 10 August 2021).
- Deng, L.J.; Qi, M.; Li, N.; Lei, Y.H.; Zhang, D.M.; Chen, J.X. Natural products, and their derivatives: Promising modulators of tumor immunotherapy. J. Leukoc. Biol. 2020, 108, 493–508. [Google Scholar] [CrossRef] [PubMed]
- Gelani, C.D.; Uy, M.M. Cytotoxicity to Artemia salina L. of marine sponge extracts from Surigao del Norte, Phillipines. Bull. Env. Pharmacol. Life Sci. 2016, 55, 14–18. [Google Scholar]
- Niksic, H.; Becic, F.; Koric, E.; Gusic, I.; Omeragic, E.; Muratovic, S.; Miladinovic, B.; Duric, K. Cytotoxicity screening of Thymus vulgaris L. essential oil in brine shrimp nauplii and cancer cell lines. Sci. Rep. 2021, 11, 13178. [Google Scholar] [CrossRef]
- Schwing, A.; Pomares, C.; Majoor, A.; Boyer, L.; Marty, P.; Michel, G. Leishmania infection: Misdiagnosis as cancer and tumor-promoting potential. Acta Trop. 2019, 1, 104855. [Google Scholar] [CrossRef]
- Khan, N.H.; ul Bari, A.; Hashim, R.; Khan, I.; Muneer, A.; Shah, A.; Wahid, S.; Yardley, V.; O’Neil, B.; Sutherland, C.J. Cutaneous leishmaniasis in Khyber Pakhtunkhwa province of Pakistan: Clinical diversity and species-level diagnosis. Am. J. Trop. Med. Hyg. 2016, 2, 1106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Kamel, M.A. Leishmaniasis and malignancy: A review and perspective. Clinic. Skin Cancer 2017, 1, 54–58. [Google Scholar] [CrossRef] [Green Version]
- Badirzadeh, A.; Heidari-Kharaji, M.; Fallah-Omrani, V.; Dabiri, H.; Araghi, A.; Salimi Chirani, A. Antileishmanial activity of Urtica dioica extract against zoonotic cutaneous leishmaniasis. PLoS Negl. Trop. Dis. 2020, 14, e0007843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, R.; Patil, S.M.; Pal, G.; Ahmad, M. Evaluation of in vivo and in vitro anti-inflammatory activity of Ajuga bracteosa Wall ex Benth. Asian Pac. J. Trop. Dis. 2012, 2, 404–407. [Google Scholar] [CrossRef]
- Hsieh, W.T.; Liu, Y.T.; Lin, W.C. Anti-inflammatory properties of Ajuga bracteosa in vivo and in vitro study and their effects on mouse model of liver fibrosis. J. Ethnopharmacol. 2011, 135, 116–125. [Google Scholar] [CrossRef]
- Ganaie, H.A.; Ali, M.N.; Ganai, B.A.; Kaur, J.; Ahmad, M. GC–MS analysis and evaluation of mutagenic and antimutagenic activity of ethyl acetate extract of Ajuga bracteosa wall ex. benth: An endemic medicinal plant of Kashmir Himalaya, India. J. Clinic. Toxicol. 2016, 6, 288. [Google Scholar] [CrossRef]
- Yousaf, T.; Rafique, S.; Wahid, F.; Rehman, S.; Nazir, A.; Rafique, J.; Aslam, K.; Shabir, G.; Shah, S.M. Phytochemical profiling and antiviral activity of Ajuga bracteosa, Ajuga parviflora, Berberis lycium and Citrus lemon against Hepatitis C Virus. Microb. Pathog. 2018, 118, 154–158. [Google Scholar] [CrossRef]
- Kayani, W.K.; Palazòn, J. Cusidò, R.M.; Mirza, B. The effect of rol genes on phytoecdysteroid biosynthesis in Ajuga bracteosa differs between transgenic plants and hairy roots. RSC Adv. 2016, 6, 22700–22708. [Google Scholar] [CrossRef]
- Mauro, M.L.; Bettini, P.P. Agrobacterium rhizogenes rolB oncogene: An intriguing player for many roles. Plant Physiol. Biochem. 2021, 165, 10–18. [Google Scholar] [CrossRef]
- Durak, T.; Depciuch, J. Effect of plant sample preparation and measuring methods on ATR-FTIR spectra results. Environ. Exp. Bot. 2020, 169, 103915. [Google Scholar] [CrossRef]
- Liu, Y.; He, Z.; Uchimiya, M. Comparison of biochar formation from various agricultural by-products using FTIR spectroscopy. Mod. Appl. Sci. 2015, 9, 246. [Google Scholar] [CrossRef]
- Bathoju, G.; Rao, K.; Giri, A. Production of sapogenins (stigmasterol and hecogenin) from genetically transformed hairy root cultures of Chlorophytum borivilianum (Safed musli). Plant Cell Tiss. Org. Cult. 2017, 131, 369–376. [Google Scholar] [CrossRef]
- Fujimoto, Y.; Nakagawa, T.; Yamada, J.; Morisaki, M. Biosynthetic origin of C-26 and C-27 of the phytoecdysteroids cyasterone and 29-norcyasterone in Ajuga hairy roots. Chem. Commun. 1996, 2063–2064. [Google Scholar] [CrossRef]
- Uozumi, N.; Ohtake, Y.; Nakashimada, Y.; Morikawa, Y.; Tanaka, N.; Kobayashi, T. Efficient regeneration from GUS-transformed Ajuga hairy root. J. Ferment. Bioeng. 1996, 81, 374–378. [Google Scholar] [CrossRef]
- Kayani, W.K.; Palazòn, J.; Cusidò, R.M.; Mirza, B. Effect of pRi T-DNA genes and elicitation on morphology and phytoecdysteroid biosynthesis in Ajuga bracteosa hairy roots. RSC Adv. 2017, 7, 47945–47953. [Google Scholar] [CrossRef] [Green Version]
- Cui, M.L.; Liu, C.; Piao, C.L.; Liu, C.L. A stable Agrobacterium rhizogenes-mediated transformation of cotton (Gossypium hirsutum L.) and plant regeneration from transformed hairy root via embryogenesis. Front. Plant Sci. 2020, 11, 604255. [Google Scholar] [CrossRef]
- Dilshad, E.; Cusido, R.M.; Estrada, K.R.; Bonfill, M.; Mirza, B. Genetic transformation of Artemisia carvifolia Buch with rol genes enhances artemisinin accumulation. PLoS ONE 2015, 10, e0140266. [Google Scholar]
- Arshad, W.; Haq, I.U.; Waheed, M.T.; Mysore, K.S.; Mirza, B. Agrobacterium-mediated transformation of tomato with rolB gene results in enhancement of fruit quality and foliar resistance against fungal pathogens. PLoS ONE 2014, 9, e96979. [Google Scholar] [CrossRef]
- Schmülling, T.; Fladung, M.; Grossmann, K.; Schell, J. Hormonal content and sensitivity of transgenic tobacco and potato plants expressing single rol genes of Agrobacterium rhizogenes T-DNA. Plant J. 1993, 3, 371–382. [Google Scholar] [CrossRef]
- Dilshad, E.; Zafar, S.; Ismail, H.; Waheed, M.T.; Cusido, R.M.; Palazon, J.; Mirza, B. Effect of rol genes on polyphenols biosynthesis in Artemisia annua and their effect on antioxidant and cytotoxic potential of the plant. Appl. Biochem. Biotechnol. 2016, 179, 1456–1468. [Google Scholar] [CrossRef]
- Kiani, B.H.; Suberu, J.; Mirza, B. Cellular engineering of Artemisia annua and Artemisia dubia with the rolABC genes for enhanced production of potent anti-malarial drug artemisinin. Malar. J. 2016, 15, 252. [Google Scholar] [CrossRef] [Green Version]
- Mauro, M.L.; Costantino, P.; Bettini, P.P. The never-ending story of rol genes: A century after. Plant Cell Tiss. Org. Cult. 2017, 131, 201–212. [Google Scholar] [CrossRef]
- Rubnawaz, S.; Kayani, W.K.; Mahmood, R.; Mirza, B. Enhanced stress tolerance in transformed Ajuga bracteosa Wall. ex Benth. regenerants by upregulated gene expression of metabolic pathways. Turk. J. Bot. 2020, 44, 410–426. [Google Scholar] [CrossRef]
- Rubnawaz, S.; Akhtar, N.; Mahmood, R.; Khan, A.; Okla, M.K.; Alamri, S.A.; Alaraidh, I.A.; Alwasel, Y.A.; Mirza, B. Polyphenol rich Ajuga bracteosa transgenic regenerants display better pharmacological potential. Molecules 2021, 26, 4874. [Google Scholar] [CrossRef]
- Ismail, H.; Dilshad, E.; Waheed, M.T.; Mirza, B. Transformation of lettuce with rolABC genes: Extracts show enhanced antioxidant, analgesic, anti-inflammatory, antidepressant, and anticoagulant activities in rats. Appl. Biochem. Biotechnol. 2017, 181, 1179–1198. [Google Scholar] [CrossRef]
- Bulgakov, V.P.; Tchernoded, G.K.; Mischenko, N.P.; Shkryl, Y.N.; Glazunov, V.P.; Fedoreyev, S.A.; Zhuravlev, Y.N. Increase in anthraquinone content in Rubia cordifolia cells transformed by rol genes does not involve activation of the NADPH oxidase signaling pathway. Biochemistry (Moscow) 2003, 68, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Shkryl, Y.N.; Veremeichik, G.N.; Bulgakov, V.P.; Tchernoded, G.K.; Mischenko, N.P.; Fedoreyev, S.A.; Zhuravlev, Y.N. Individual and combined effects of the rolA, B, and C genes on anthraquinone production in Rubia cordifolia transformed calli. Biotechnol. Bioeng. 2008, 100, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Bulgakov, V.P.; Khodakovskaya, M.V.; Labetskaya, N.V.; Chernoded, G.K.; Zhuravlev, Y.N. The impact of plant rolC oncogene on ginsenoside production by ginseng hairy root cultures. Phytochemistry 1998, 49, 1929–1934. [Google Scholar] [CrossRef]
- Dubrovina, A.S.; Manyakhin, A.Y.; Zhuravlev, Y.N.; Kiselev, K.V. Resveratrol content and expression of phenylalanine ammonia-lyase and stilbene synthase genes in rolC transgenic cell cultures of Vitis amurensis. Appl. Microbiol. Biotechnol. 2010, 88, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Palazón, J.; Cusidó, R.M.; Roig, C.; Pinol, M.T. Expression of the rolC gene and nicotine production in transgenic roots and their regenerated plants. Plant Cell Rep. 1998, 17, 384–390. [Google Scholar]
- Abbasi, M.S.; Tahir, M.A.; Meer, S. FTIR Spectroscopic study of aloe vera barbadensis Mill Buds. Asian J. Chem. Sci. 2020, 7, 1–6. [Google Scholar] [CrossRef]
- dos Santos Grasel, F.; Ferrão, M.F.; Wolf, C.R. Development of methodology for identification the nature of the polyphenolic extracts by FTIR associated with multivariate analysis. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2016, 153, 94–101. [Google Scholar] [CrossRef]
- Donadu, M.G.; Peralta-Ruiz, Y.; Usai, D.; Maggio, F.; Molina-Hernandez, J.B.; Rizzo, D.; Bussu, F.; Rubino, S.; Zanetti, S.; Paparella, A.; et al. Colombian essential oil of Ruta graveolens against nosocomial antifungal resistant Candida strains. J. Fungus. 2021, 7, 383. [Google Scholar] [CrossRef]
- Langeveld, W.T.; Veldhuizen, E.J.; Burt, S.A. Synergy between essential oil components and antibiotics: A review. Crit. Rev. Microbiol. 2014, 40, 76–94. [Google Scholar] [CrossRef] [PubMed]
- Brochot, A.; Guilbot, A.; Haddioui, L.; Roques, C. Antibacterial, antifungal, and antiviral effects of three essential oil blends. MicrobiologyOpen 2017, 6, e00459. [Google Scholar] [CrossRef] [PubMed]
- Ganaie, H.A.; Ali, M.N.; Ganai, B.A.; Meraj, M.; Ahmad, M. Antibacterial activity of 14, 15-dihydroajugapitin and 8-o-acetylharpagide isolated from Ajuga bracteosa Wall ex. Benth against human pathogenic bacteria. Microb. Pathog. 2017, 103, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Zahra, S.S.; Ahmed, M.; Qasim, M.; Gul, B.; Zia, M.; Mirza, B.; Haq, I.U. Polarity based characterization of biologically active extracts of Ajuga bracteosa Wall. ex Benth and RP-HPLC analysis. BMC Complement. Altern. Med. 2017, 17, 443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rehman, N.U.; Begum, N.; Ali, L.; Al-Harrasi, A.; Abbas, G.; Ahmad, S.; Khan, A.L.; Shinwari, Z.K.; Hussain, J. Lipid peroxidation, antiglycation, cytotoxic, phytotoxic, antioxidant, antiplatelet and antimicrobial activities of Ajuga bracteosa against various pathogens. Pak. J. Bot. 2015, 47, 1195–1197. [Google Scholar]
- Vohra, A.; Kaur, H. Chemical investigation of medicinal plant Ajuga bracteosa. J. Nat. Prod. Plant. Resour. 2011, 1, 37–45. [Google Scholar]
- Loganayaki, N.; Siddhuraju, P.; Manian, S. Antioxidant activity and free radical scavenging capacity of phenolic extracts from Helicteres isora L. and Ceiba pentandra L. J. Food Sci. Technol. 2013, 50, 687–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shabbir, M.; Khan, M.R.; Saeed, N. Assessment of phytochemicals, antioxidant, anti-lipid peroxidation and anti-hemolytic activity of extract and various fractions of Maytenus royleanus leaves. BMC Complement. Altern. Med. 2013, 13, 143. [Google Scholar] [CrossRef] [Green Version]
- Imran, M.; Jan, H.; Faisal, S.; Shah, S.A.; Shah, S.; Khan, M.N.; Akbar, M.T.; Rizwan, M.; Jan, F.; Syed, S. In vitro examination of anti-parasitic, anti-Alzheimer, insecticidal and cytotoxic potential of Ajuga bracteosa Wallich leaves extracts. Saudi J. Biol. Sci. 2021, 28, 3031–3036. [Google Scholar] [CrossRef] [PubMed]
- Solowey, E.; Lichtenstein, M.; Sallon, S.; Paavilainen, H.; Solowey, E.; Lorberboum-Galski, H. Evaluating medicinal plants for anticancer activity. Sci. World J. 2014, 2014, 721402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pal, A.; Toppo, F.A.; Chaurasiya, P.K.; Singour, P.K.; Pawar, R.S. In vitro cytotoxicity study of methanolic fraction from Ajuga bracteosa wall ex. benth on MCF-7 breast adenocarcinoma and hep-2 larynx carcinoma cell lines. Pharmacogn. Res. 2014, 6, 87. [Google Scholar]
- Hussain, M.; Bibi, Y.; Raja, N.I.; Iqbal, M.; Aslam, S.; Tahir, N.; Imran, M.; Iftikhar, A. A review of therapeutic potential of Ajuga bracteosa: A critically endangered plant from Himalaya. J. Coast. Life Med. 2016, 4, 918–924. [Google Scholar]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Preparation of plasmid DNA by alkaline lysis with SDS: Minipreparation. Cold Spring Harb. Protoc. 2006, 2006, pdb.prot4084. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Jha, S. Effects associated with insertion of rol genes on morphogenic potential in explants derived from transgenic Bacopa monnieri (L.) Wettst. Plant Cell Tiss. Org. Cult. 2021, 12, 1–2. [Google Scholar]
- Luz, A.C.; Pretti, I.R.; Batitucci, M. Comparison of RNA extraction methods for Passiflora edulis sims leaves. Rev. Bras. Frutic. 2016, 38, 226–232. [Google Scholar] [CrossRef] [Green Version]
- Veremeichik, G.N.; Bulgakov, V.P.; Shkryl, Y.N.; Silantieva, S.A.; Makhazen, D.S.; Tchernoded, G.K.; Mischenko, N.P.; Fedoreyev, S.A.; Vasileva, E.A. Activation of anthraquinone biosynthesis in long-cultured callus culture of Rubia cordifolia transformed with the rolA plant oncogene. J. Biotechnol. 2019, 306, 38–46. [Google Scholar] [CrossRef]
- Meziti, H.; Bouriche, H.; Kada, S.; Demirtas, I.; Kizil, M.; Senator, A.; Garrido, G. Phytochemical analysis, and antioxidant, anti-hemolytic and genoprotective effects of Quercus ilex L. and Pinus halepensis Mill. methanolic extracts. J. Pharm. Pharmacogn. Res. 2019, 7, 260–272. [Google Scholar]
- McLaughlin, J.L.; Rogers, L.L.; Anderson, J.E. The use of biological assays to evaluate botanicals. Drug Inf. J. 1998, 32, 513–524. [Google Scholar] [CrossRef]
- Bhukya, B.R.; Yellu, N.R. Evaluation of anticancer activity of methanolic extract of Hiptage benghalensis (L.) Kurz on cancer cell lines. Pharmacogn. Res. 2018, 10, 309–313. [Google Scholar]
- Ahmad, B.; Islam, A.; Khan, A.; Khan, M.A.; ul Haq, I.; Jafri, L.; Ahmad, M.; Mehwish, S.; Khan, A.; Ullah, N. Comprehensive investigations on anti-leishmanial potentials of Euphorbia wallichii root extract and its effects on membrane permeability and apoptosis. Comp. Immunol. Microbiol. Infect. Dis. 2019, 64, 138–145. [Google Scholar] [CrossRef] [PubMed]





| No. | Functional Groups of Active Components in Crude Extracts | |||||||
|---|---|---|---|---|---|---|---|---|
| WT | ABRL1 | ABRL2 | ABRL3 | |||||
| Functional Groups | Peak Value | Functional Groups | Peak Value | Functional Groups | Peak Value | Functional Groups | Peak Value | |
| 1 | Primary/Secondary amines | 3287.70 | Primary/Secondary amines | 3270.74 | Primary/Secondary amines | 3272.71 | Primary/Secondary amines | 3283.41 |
| 2 | Alkane | 2918.21 | Alkane | 2917.51 | Alkane | 2917.72 | Alkane | 2918.39 |
| 3 | Carboxylic acid, alkane | 2850.07 | Carboxylic acid, alkane | 2849.59 | Carboxylic acid, alkane | 2849.76 | Carboxylic acid, alkane | 2850.21 |
| 4 | Amino acid | 1631.70 | Ketone, carboxylic acid | 1710.23 | Ketone, carboxylic acid | 1708.47 | Thiol/mercaptan | 2349.30 |
| 5 | Alkane, Nitro compounds | 1376.57 | Amino acid | 1594.74 | Amino acid | 1623.07 | Amino acid | 1625.01 |
| 6 | Fluoride, alkyl halide | 1248.95 | Aromatic | 1414.52 | Alkane, Nitro compounds | 1375.99 | Alkane | 1406.24 |
| 7 | Alcohol, aliphatic amines | 1029.22 | Alkane, Nitro compounds | 1373.30 | Fluoride, alkyl halide, aliphatic amines | 1241.37 | Fluoride, alkyl halide | 1249.00 |
| 8 | p-substituted alcohols/phenols | 815.80 | Fluoride, alkyl halide, aliphatic amines | 1237.64 | Ether | 1148.72 | Ether | 1147.89 |
| 9 | Aromatic compound, primary/secondary amines | 776.54 | Alcohol, aliphatic amines | 1014.17 | Alcohol, aliphatic amines | 1026.78 | Alcohol, aliphatic amines | 1026.83 |
| 10 | Alkynes, alkyl halide, Aryl disulfide | 518.90 | p-substituted alcohols/phenols | 812.42 | p-substituted alcohols/phenols | 815.76 | Primary, secondary amines | 866.90 |
| 11 | Alkynes, alkyl halide, Aryl disulfide | 525.19 | Aromatic compound, primary/secondary amines | 776.27 | p-substituted alcohols/phenols | 815.63 | ||
| 12 | Alkynes, alkyl halide, Aryl disulfide | 520.25 | Aromatic compound, primary/secondary amines | 776.14 | ||||
| 13 | Alkynes, alkyl halide, Aryl disulfide | 517.93 | ||||||
| Samples | Zone of Inhibition at 100 µg/Disc (mm) | |||||||
|---|---|---|---|---|---|---|---|---|
| M. luteus | MIC (µg/mL) | S. aureus | MIC (µg/mL) | E. aerogenes | MIC (µg/mL) | E. coli | MIC (µg/mL) | |
| WT | 7.3 ± 0.2 e | - | 6.9 ± 0.3 f | - | 6.5 ± 0.1 f | - | 7.1 ± 0.4 f | - |
| ABRL1 | 13.7 ± 0.6 c | >100 | 15.1 ± 0.5 b,c | >100 | 8.3 ± 0.2 d | - | 13.9 ± 0.4 c | >100 |
| ABRL2 | 13.8 ± 0.5 c | >100 | 15.5 ± 0.4 b | >100 | 8.9 ± 0.4 d | - | 14.5 ± 0.3 c | >100 |
| ABRL3 | 16.1 ± 0.6 b | >100 | 16.9 ± 0.7 b | >100 | 8.2 ± 0.3 d | - | 16.2 ± 0.5 b | >100 |
| kanamycin | 26.2 ± 1 a | 0.31 | 25.1 ± 2 a | 0.33 | 23.7 ± 1 a | 0.29 | 24.4 ± 3 a | 0.30 |
| Samples | % Hemolysis | CC50 (µg/mL) | ||
|---|---|---|---|---|
| Test Concentrations (µg/mL) | ||||
| 1000 | 500 | 250 | ||
| WT | 16.25 ± 2 b | 5.25 ± 0.9 c | 2.97 ± 1 d | 1952.63 ± 12 |
| ABRL1 | 9.76 ± 1 b,c | 4.01 ± 5 c,d | 0.82 ± 0.07 e | 5309.04 ± 13 |
| ABRL2 | 8.43 ± 0.8 b,c | 2.87 ± 6 d | 2.72 ± 0.3 d | 6179.54 ± 9 |
| ABRL3 | 6.28 ± 0.3 c | 2.41 ± 4 d | 0.88 ± 0.1 e | 7293.05 ± 7 |
| AAPH | 100 a | 0.07 | ||
| Samples | % Mortality After 24 h | LD50 (µg/mL) | ||
|---|---|---|---|---|
| Test Concentrations (µg/mL) | ||||
| 200 | 66 | 21.8 | ||
| WT | 70.33 ± 3 b | 50.4 ± 2 d | 30.3 ± 3 f | 75.6 ± 9 |
| ABRL1 | 79.3 ± 5 a | 60.3 ± 2 c | 40.0 ± 3 e | 39.6 ± 4 |
| ABRL2 | 68.6 ± 4 b | 58.6 ± 1 c | 40.6 ± 2 e | 41.62 ± 2 |
| ABRL3 | 67.14 ± 5 b | 59.2 ± 3 c | 42.3 ± 4 e | 43.62 ± 5 |
| Doxorubicin | 84.65 ± 10 a | 5.8 ± 0.3 | ||
| Treatment | IC50 (µg/mL) | |||||
|---|---|---|---|---|---|---|
| HepG2 | LM3 | A549 | HT-29 | MCF-7 | MDA-MB-231 | |
| WT | >100 | >100 | >100 | >100 | >100 | >100 |
| ABRL1 | 71.2 ± 3.1 d | 58.3 ± 2.3 c | 98.1 ± 2.4 f | 98.2 ± 3.3 f | >100 | >100 |
| ABRL2 | 79.5 ± 4.6 d,e | 53.4 ± 1.5 c | 94.2 ± 1.7 f | 99.7 ± 1.9 f | >100 | >100 |
| ABRL3 | 57.1 ± 2.2 c | 46.2 ± 1.1 b | 72.4 ± 1.3 d | 73.3 ± 2.1 d | 98.7 ± 1.6 f | 97.1 ± 2.5 f |
| Ellipticine | 0.38 ± 0.01 a | 0.47 ± 0.04 a | 0.41 ± 0.05 a | 0.39 ± 0.04 a | 0.35 ± 0.02 a | 0.45 ± 0.03 a |
| Samples | % Mortality after 72 h | IC50 (µg/mL) | ||
|---|---|---|---|---|
| Test Concentrations (µg/mL) | ||||
| 1000 | 500 | 250 | ||
| W | 63.52 ± 7 d | 55.64 ± 4 e | 47.23 ± 2 f | 313.99 ± 6 |
| ABRL1 | 78.14 ± 1 b | 67.38 ± 1 c | 56.63 ± 5 e | 163.04 ± 8 |
| ABRL2 | 75.48 ± 6 b | 68.42 ± 6 c, | 61.69 ± 6 d | 77.53 ± 7 |
| ABRL3 | 74.53 ± 8 b | 69.48 ± 4 c, | 62.14 ± 3 d | 56.16 ± 2 |
| Amphotericin B | 100 a | 0.01 | ||
| Genes | GenBank Accession No. | Primer Sequences | Size (bp) |
|---|---|---|---|
| House Keeping Genes | |||
| βActin | DQ531565 | F: GATTGAGCACGGTATTGTTAG R: ACACCATCACCAGAATCCAAC | 259 |
| 18s | HQ730915.1 | F: GGAGAGGGAGCCTGAGAAAC R: GATTTAGATTGTACTCATTCC | 122 |
| Agrobacterium rhizogenes genes | |||
| rolB (qPCR) | X03433 | F: CGAGGGACTGAAAACCGCC R: CCGAGAGTCGCAGGGTTAG | 127 |
| rolB (Simple PCR) | X15952.1 | F: GCTCTTGCAGTGCTAGATTT R: GAAGGTGCAAGCTACCTCTC | 779 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rubnawaz, S.; Okla, M.K.; Akhtar, N.; Khan, I.U.; Bhatti, M.Z.; Duong, H.-Q.; El-Tayeb, M.A.; Elbadawi, Y.B.; Almaary, K.S.; Moussa, I.M.; et al. Antibacterial, Antihemolytic, Cytotoxic, Anticancer, and Antileishmanial Effects of Ajuga bracteosa Transgenic Plants. Plants 2021, 10, 1894. https://doi.org/10.3390/plants10091894
Rubnawaz S, Okla MK, Akhtar N, Khan IU, Bhatti MZ, Duong H-Q, El-Tayeb MA, Elbadawi YB, Almaary KS, Moussa IM, et al. Antibacterial, Antihemolytic, Cytotoxic, Anticancer, and Antileishmanial Effects of Ajuga bracteosa Transgenic Plants. Plants. 2021; 10(9):1894. https://doi.org/10.3390/plants10091894
Chicago/Turabian StyleRubnawaz, Samina, Mohammad K. Okla, Nosheen Akhtar, Imdad Ullah Khan, Muhammad Zeeshan Bhatti, Hong-Quan Duong, Mohamed A. El-Tayeb, Yahaya B. Elbadawi, Khalid S. Almaary, Ihab M. Moussa, and et al. 2021. "Antibacterial, Antihemolytic, Cytotoxic, Anticancer, and Antileishmanial Effects of Ajuga bracteosa Transgenic Plants" Plants 10, no. 9: 1894. https://doi.org/10.3390/plants10091894
APA StyleRubnawaz, S., Okla, M. K., Akhtar, N., Khan, I. U., Bhatti, M. Z., Duong, H. -Q., El-Tayeb, M. A., Elbadawi, Y. B., Almaary, K. S., Moussa, I. M., Abbas, Z. K., & Mirza, B. (2021). Antibacterial, Antihemolytic, Cytotoxic, Anticancer, and Antileishmanial Effects of Ajuga bracteosa Transgenic Plants. Plants, 10(9), 1894. https://doi.org/10.3390/plants10091894