Allelopathic Potential of Mangroves from the Red River Estuary against the Rice Weed Echinochloa crus-galli and Variation in Their Leaf Metabolome
Abstract
:1. Introduction
2. Results
2.1. Comparison of Source Species Chemical Fingerprints
2.2. Relative Allelopathic Effects
2.2.1. Effects on Seed Germination
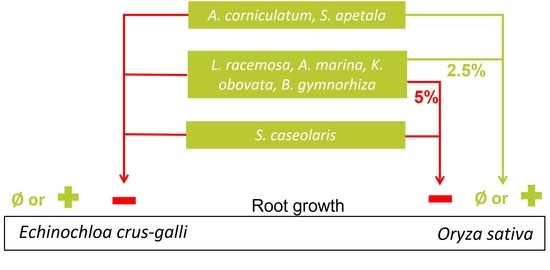
2.2.2. Effects on Seedling Growth
2.2.3. Selection of Candidate Species for Further Chemical Investigation
2.3. A. corniculatum and S. apetala Biomarker Selection and Annotation
| Biomarker | [M-H]− m/z | Adduct | RT (min) | Molecular Formula | Mass Error (ppm) | Mσ a | MS/MS Fragment Ions (Relative Abundance in %) | Putative Identification | Reference | |
|---|---|---|---|---|---|---|---|---|---|---|
| Aegiceras corniculatum | M943T476 | 943.4872 | 979.4685 [M+Cl]− 1011.4771 [M-H+HCOONa]− | 7.94 | C47H76O19 | 3.8 | 36.7 | 943.4872 [M-H]− (38.4), 781.4355 [M-H-Hexose]− (100), 619.3829 [M-H-2Hexose]− (63.2), 487.3426 [M-H-2Hexose-Pentose]− [C30H47O5]− (38.5), 275.0768 (42.2), 113.0243 (28.7), 101.0239 (33.8), 89.0238 (29.3) | (3β, 16α, 20α)-3,16,28-trihydroxyolean-12-en-29-oic acid 3-{O-β-D-glucopyranosyl (1→2)-O-[β-D-glucopyranosyl (1→4)]-α-L-arabinopyranoside} | [41] |
| M457T495 | 457.1700 | 479.1545 [M-2H+Na]− 523.1408 [M-H+HCOONa]− | 8.25 | C21H30O11 (or C20H31N2O8P) | −0.3 4.9 | 30.6 8.9 | 238.0854 (100.0), 235.1335 (2.7), 235.0973 (4.8), 220.0748 (5.3), 194.0945 (11.3), 192.0783 (3.1), 177.0913 (3.0), 165.0553 (5.5), 151.0393 (3.3) | Unknown | ||
| M781T518 | 781.4384 | 817.4162 [M+Cl]− 849.4374 [M-H+HCOONa]− | 8.64 | C41H66O14 | 0.8 | 7.0 | 781.4384 [M-H]− (31.7), 619.3849 [M-H-Hexose]− (100), 601.3749 (22.7), 487.3456 [M-H-Hexose-Pentose]− [C30H47O5]− (10.3), 101.0242 (19.3), 89.0244 (18.7) | Triterpenoid saponin | [41] | |
| M410T546 | 409.1175 | 477.1044 [M-H+HCOONa]− | 9.10 | C16H26O10S | −2.3 | 9.5 | 241.0029 [C6H9O8S]− (9.1), 138.9708 [C2H3O5S]− (4.4), 96.9600 [HSO4]− (100) | Monoterpene sulfate | ||
| Sonneratia apetala | M895T343 Sa M1 | 447.0941 | 505.0515 [M+NaCl-H]− 515.0801 [M-H+HCOONa]− | 5.72 | C21H20O11 | −1.8 | 16.5 | 357.0624 (22.9), 339.0514 (21.3), 327.0516 (79.4), 311.0565 (26), 298.0483 (100), 285.0405 [M-H-Hexose]− [C15H9O6]− (51.4), 284.033 [M-H-Hexose]− (38.1), 199.0407 (5.4), 175.0405 (8.6), 151.0039 (3.4), 133.0298 (36.7) | Luteolin 7-O-β-glucoside | [46,47,48,49] |
| M607T418 Sa M4 | 607.1671 | 643.1442 [M+Cl]− 675.1545 [M+HCOONa-H]− | 6.97 | C28H32O15 | −0.2 | 5.0 | 299.0563 [M-H-rutinose]− (53.5), 284.0329 [M-H-rutinose-CH3]− (100) | Diosmin a | [44,45] | |
| M481T560 Sa M6 | 423.0023 | 480.9607 [M+NaCl-H]− 490.9817 [M+HCOONa-H]− | 9.34 | C17H12O11S | 0.2 | 10.8 | 343.0471 [M-H-SO3]− (0.3), 328.0227 [M-H-SO3-CH3]− (20.6), 312.9992 [M-H-SO3-2CH3]− (100), 297.9757 [M-H-SO3-3CH3]− (51.4), 285.0042 (17.2), 269.9807 (8.8) | 3,3′,4′-trimethylellagic acid 4-sulfate | [40] |
| Compound | [M-H]− m/z | Adduct | RT (min) | Molecular Formula | Mass Error (ppm) | Mσ a | MS/MS m/z Fragment Ions (Relative Abundance in %) | Putative Identification | Reference | |
|---|---|---|---|---|---|---|---|---|---|---|
| Aegiceras corniculatum | Ac M1 | 493.0607 | 515.0438 [M+Na-2H]− | 5.75 | C21H18O14 | 3.3 | 23.1 | 317.0291 [M-H-glucuronic acid]− (62.3), 271.0237 (29.1), 261.0395 (12.5), 243.0289 (11.0), 178.9980 (56.8), 163.0030 (11.4), 151.0031 (100), 137.0239 (53.2), 109.0291 (12.9), 107.0134 (17.6) | Myricetin-3-glucuronide | [42,43,52] |
| Ac M2 | 477.067 | 499.0502 [M+Na-2H]− | 6.37 | C21H18O13 | 1.0 | 21.4 | 301.0350 [M-H-glucuronic acid]− (72.3), 283.0242 (17.0), 255.0295 (26), 245.0451 (27.3), 178.9983 (32.4), 163.0033 (20.7), 151.0034 (100), 121.0293 (30.3), 109.0292 (19.7), 107.0136 (23.2) | Quercetin glucuronide | [43] | |
| Ac M3 | 447.0924 | 483.0692 [M+Cl]− 515.0804 [M+HCOONa-H]− | 6.90 | C21H20O11 | 2.0 | 15.2 | 300.0273 (6.6), 284.0323 (38.7), 271.0242 (10.0), 255.0296 (100), 227.0347 (85.0) | Kaempferol-3-O-glucoside a (Astragalin) | [41] | |
| Ac M4 | 451.1638 | 519.1499 [M+HCOONa-H]− | 7.08 | C19H32O10S | 1.2 | 22.5 | 451.1636 [C19H31O10S]− (25.8), 256.9969 [C6H9O9S]− (6.3), 241.0015 [C6H9O8S]− (6.5), 177.0401 [C6H9O6]− (2.8), 138.9704 [C2H3O5S]− (2.2), 96.9597 [HSO4]− (100), 79.9571 [SO3]− (3.5) | Unknown | ||
| Sonneratia apetala | Sa M1 | |||||||||
| Sa M2 | 787.1008 | 809.0818 [M+Na-2H]− 823.0780 [M+Cl]− | 6.00 | C34H27O22 | −1.1 | 9.4 | 635.0915 (0.3), 483.0789 (1.1), 465.0678 (50.2), 313.0569 (47.7), 295.0463 (18.8), 169.0145 (100), 125.0247 (26.8) | Tetragalloyl glucose | [51,53,54] | |
| Sa M3 | 431.0998 | 499.0861 [M+HCOONa-H]− | 6.25 | C21H20O10 | −3.4 | 9.6 | 323.0566 (13.1), 311.0569 (31.2), 295.0618 (11.3), 283.0617 (100), 281.0460 (20.6), 269.0457 (11.1), 117.0350 (21.6) | Vitexin a | [50] | |
| Sa M4 | ||||||||||
| Sa M5 | 408.9880 | 329.0310 [M-SO3-H]− 476.9758 [M+HCOONa-H]− | 7.82 | C16H10O11S | −2.1 | 20.9 | 329.0303 [M-H-SO3]− (0.6), 314.0067 [M-H-SO3-CH3]− (37.0), 298.9834 [C14H3O8]− [M-H-SO3-2CH3]− (100), 270.9882 [C13H3O7]− (48.9), 242.9934 [C12H3O6]− (2.6) | Dimethyl ellagic acid sulfate | [40] | |
| Sa M6 |
3. Discussion
3.1. Leaf Chemical Fingerprint Similarities According to Phylogenetic Proximity
3.2. Contrasted Allelopathic Effects Depending on Mangrove Species
3.2.1. Allelopathic Effects on Germination
3.2.2. Allelopathic Effects on Root Growth
3.2.3. A. corniculatum and S. apetala Are Interesting Candidates for Further Investigations
3.3. A. corniculatum and S. apetala Chemical Investigation
3.3.1. A. corniculatum Biomarkers and Compounds as Potential Allelochemicals
3.3.2. S. apetala Biomarkers and Compounds as Potential Allelochemicals
3.3.3. First Description of Sulfated Phenolics in S. apetala
3.4. Perspectives for Further Field Assays
4. Materials and Methods
4.1. Sampling Site
4.2. Material Collection
- -
- Three species belonging to the Rhizophoraceae family: Rhizophora stylosa Griff, Bruguiera gymnorhiza Buch ham and Kandelia obovata (L.) Lam.
- -
- Two from the Sonneratia genus (Lythraceae): S. caseolaris (L.) Engl. and S. apetala Sheue, Liu & Yong sp.nov
- -
- Three other species belonging to different families: Aegiceras corniculatum (L.) Blanco (Primulaceae), Lumnitzera racemosa Willd (Combretaceae) and Avicennia marina (Forssk.) Vierh (Acanthaceae).
4.3. Allelopathy Bioassay
4.4. Germination Parameters
4.5. Growth Parameters
4.6. Metabolomic Analysis, Data Pre-Processing and Annotation of Metabolites
4.7. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lundkvist, A.; Verwijst, T. Weed Biology and Weed Management in Organic Farming. In Research in Organic Farming; IntechOpen: London, UK, 2011. [Google Scholar]
- Vigueira, C.C.; Olsen, K.M.; Caicedo, A.L. The Red Queen in the Corn: Agricultural Weeds as Models of Rapid Adaptive Evolution. Heredity 2013, 110, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Abbas, T.; Zahir, Z.A.; Naveed, M.; Kremer, R.J. Chapter Five—Limitations of Existing Weed Control Practices Necessitate Development of Alternative Techniques Based on Biological Approaches. In Advances in Agronomy; Sparks, D.L., Ed.; Academic Press: Cambridge, MA, USA, 2018; Volume 147, pp. 239–280. [Google Scholar]
- Datta, A.; Ullah, H.; Ferdous, Z. Water Management in Rice. In Rice Production Worldwide; Chauhan, B.S., Jabran, K., Mahajan, G., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 255–277. [Google Scholar]
- Cosslett, T.L.; Cosslett, P.D. Rice Cultivation, Production, and Consumption in Mainland Southeast Asian Countries: Cambodia, Laos, Thailand, and Vietnam. In Sustainable Development of Rice and Water Resources in Mainland Southeast Asia and Mekong River Basin; Cosslett, T.L., Cosslett, P.D., Eds.; Springer: Singapore, 2018; pp. 29–53. [Google Scholar]
- Maitah, K.; Smutka, L.; Sahatqija, J.; Maitah, M.; Phuong Anh, N. Rice as a Determinant of Vietnamese Economic Sustainability. Sustainability 2020, 12, 5123. [Google Scholar] [CrossRef]
- Kraehmer, H.; Jabran, K.; Mennan, H.; Chauhan, B.S. Global Distribution of Rice Weeds—A Review. Crop Prot. 2016, 80, 73–86. [Google Scholar] [CrossRef]
- Barrett, S.H. Crop Mimicry in Weeds. Econ. Bot. 1983, 37, 255–282. [Google Scholar] [CrossRef]
- Norsworthy, J.K.; Wilson, M.J.; Scott, R.C.; Gbur, E.E. Herbicidal Activity on Acetolactate Synthase-Resistant Barnyardgrass (Echinochloa crus-galli) in Arkansas, USA. Weed Biol. Manag. 2014, 14, 50–58. [Google Scholar] [CrossRef]
- Oerke, E.-C.; Dehne, H.-W. Safeguarding Production—Losses in Major Crops and the Role of Crop Protection. Crop Prot. 2004, 23, 275–285. [Google Scholar] [CrossRef]
- Macías, F.A.; Mejías, F.J.; Molinillo, J.M. Recent Advances in Allelopathy for Weed Control: From Knowledge to Applications. Pest Manag. Sci. 2019, 75, 2413–2436. [Google Scholar] [CrossRef]
- Rice, E.L. Allelopathy, 2nd ed.; Academic Press: New York, NY, USA, 1984. [Google Scholar]
- Inderjit; Duke, S.O. Ecophysiological Aspects of Allelopathy. Planta 2003, 217, 529–539. [Google Scholar] [CrossRef]
- Wardle, D.A.; Nilsson, M.-C.; Gallet, C.; Zackrisson, O. An Ecosystem-Level Perspective of Allelopathy. Biol. Rev. 1998, 73, 305–319. [Google Scholar] [CrossRef]
- Cheng, F.; Cheng, Z. Research Progress on the Use of Plant Allelopathy in Agriculture and the Physiological and Ecological Mechanisms of Allelopathy. Front. Plant Sci. 2015, 6, 1020. [Google Scholar] [CrossRef] [Green Version]
- Vivanco, J.M.; Bais, H.P.; Stermitz, F.R.; Thelen, G.C.; Callaway, R.M. Biogeographical Variation in Community Response to Root Allelochemistry: Novel Weapons and Exotic Invasion. Ecol. Lett. 2004, 7, 285–292. [Google Scholar] [CrossRef]
- Fernandez, C.; Santonja, M.; Gros, R.; Monnier, Y.; Chomel, M.; Baldy, V.; Bousquet-Mélou, A. Allelochemicals of Pinus halepensis as Drivers of Biodiversity in Mediterranean Open Mosaic Habitats During the Colonization Stage of Secondary Succession. J. Chem. Ecol. 2013, 39, 298–311. [Google Scholar] [CrossRef] [PubMed]
- Linhart, Y.B.; Gauthier, P.; Keefover-Ring, K.; Thompson, J.D. Variable Phytotoxic Effects of Thymus vulgaris (Lamiaceae) Terpenes on Associated Species. Int. J. Plant Sci. 2015, 176, 20–30. [Google Scholar] [CrossRef]
- Gavinet, J.; Santonja, M.; Baldy, V.; Hashoum, H.; Peano, S.; Tchong, T.; Gros, R.; Greff, S.; Fernandez, C.; Bousquet-Mélou, A. Phenolics of the Understory Shrub Cotinus coggygria Influence Mediterranean Oak Forests Diversity and Dynamics. For. Ecol. Manag. 2019, 441, 262–270. [Google Scholar] [CrossRef]
- Hashoum, H.; Santonja, M.; Gauquelin, T.; Saatkamp, A.; Gavinet, J.; Greff, S.; Lecareux, C.; Fernandez, C.; Bousquet-Mélou, A. Biotic Interactions in a Mediterranean Oak Forest: Role of Allelopathy along Phenological Development of Woody Species. Eur. J. Forest Res. 2017, 136, 699–710. [Google Scholar] [CrossRef]
- Muzell Trezzi, M.; Vidal, R.A.; Balbinot Junior, A.A.; von Hertwig Bittencourt, H.; da Silva Souza Filho, A.P. Allelopathy: Driving Mechanisms Governing Its Activity in Agriculture. J. Plant Interact. 2016, 11, 53–60. [Google Scholar] [CrossRef]
- Jabran, K.; Mahajan, G.; Sardana, V.; Chauhan, B.S. Allelopathy for Weed Control in Agricultural Systems. Crop Prot. 2015, 72, 57–65. [Google Scholar] [CrossRef]
- Khanh, T.D.; Xuan, T.D.; Chung, I.M. Rice Allelopathy and the Possibility for Weed Management. Ann. Appl. Biol. 2007, 151, 325–339. [Google Scholar] [CrossRef]
- Khanh, T.D.; Cong, L.C.; Chung, I.M.; Xuan, T.D.; Tawata, S. Variation of Weed-Suppressing Potential of Vietnamese Rice Cultivars against Barnyardgrass (Echinochloa crus-galli) in Laboratory, Greenhouse and Field Screenings. J. Plant Interact. 2009, 4, 209–218. [Google Scholar] [CrossRef]
- Berendji, S.; Asghari, J.B.; Matin, A.A. Allelopathic Potential of Rice (Oryza sativa) Varieties on Seedling Growth of Barnyardgrass (Echinochloa crus-galli). J. Plant Interact. 2008, 3, 175–180. [Google Scholar] [CrossRef]
- Fromard, F.; Puig, H.; Mougin, E.; Marty, G.; Betoulle, J.L.; Cadamuro, L. Structure, Above-Ground Biomass and Dynamics of Mangrove Ecosystems: New Data from French Guiana. Oecologia 1998, 115, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Duke, N. Mangrove Floristics and Biogeography Revisited: Further Deductions from Biodiversity Hot Spots, Ancestral Discontinuities, and Common Evolutionary Processes. In Mangrove Ecosystems: A Global Biogeographic Perspective: Structure, Function, and Services; Springer Science+ Business Media: Berlin, Germany, 2017; pp. 17–53. ISBN 978-3-319-62204-0. [Google Scholar]
- Friess, D.; Yando, E.; Alemu, I.J.; Wong, L.-W.; Soto, S.; Bhatia, N. Ecosystem Services and Disservices of Mangrove Forests and Salt Marshes. Oceanogr. Mar. Biol. 2020, 58, 107–142. [Google Scholar]
- Pedrol, N.; González, L.; Reigosa Roger, M. Allelopathy and Abiotic Stress. In Allelopathy: A Physiological Process with Ecological Implications; Springer Science+ Business Media: Berlin, Germany, 2006; pp. 171–209. ISBN 978-1-4020-4279-9. [Google Scholar]
- Chen, L.; Peng, S. Allelopathic Potential of Mangrove Plants (Avicennia marina, Aegiceras corniculata and Bruguiera gymnorrhiza). Allelopathy J. 2008, 22, 213–220. [Google Scholar]
- Chen, L.; Peng, S.L.; Chen, B.-M.; Li, J.; Pang, J. Effects of Aqueous Extracts of 5 Mangrove Spp. on Cabbage Germination and Hypocotyl Growth of Kandelia candel. Allelopathy J. 2009, 23, 469–476. [Google Scholar]
- Jing, L.; ShaoLin, P.; Chen, L.; Wang, R.; Ni, G. Use of Sonneratia apetala Allelopathy to Control Spartina alterniflora Weed. Allelopathy J. 2010, 25, 123–132. [Google Scholar]
- Othman, R.; Ramya, R.; Mohd Hassan, N.; Daud, W.; Johari, N.N. Characterisation of Allelochemical Compounds Signature in Two Mangrove Forest Species of Rhizophora apiculata and Acrostichum aureum and Potential in Suppressing Weed Growth. IOP Conf. Ser. Earth Environ. Sci. 2019, 380, 012016. [Google Scholar] [CrossRef]
- Wolfender, J.-L.; Marti, G.; Thomas, A.; Bertrand, S. Current Approaches and Challenges for the Metabolite Profiling of Complex Natural Extracts. J. Chromatogr. A 2015, 1382, 136–164. [Google Scholar] [CrossRef]
- Favre, L.; Ortalo-Magné, A.; Greff, S.; Pérez, T.; Thomas, O.P.; Martin, J.-C.; Culioli, G. Discrimination of Four Marine Biofilm-Forming Bacteria by LC–MS Metabolomics and Influence of Culture Parameters. J. Proteome Res. 2017, 16, 1962–1975. [Google Scholar] [CrossRef]
- Paix, B.; Othmani, A.; Debroas, D.; Culioli, G.; Briand, J.-F. Temporal Covariation of Epibacterial Community and Surface Metabolome in the Mediterranean Seaweed Holobiont Taonia atomaria. Environ. Microbiol. 2019, 21, 3346–3363. [Google Scholar] [CrossRef]
- Bidve, S.C. Metabolite Profiling and Principle Component Analysis of a Mangrove Plant Aegiceras corniculatum L. (Blanco). Int. J. Pharm. Pharmacol. 2018, 2, 1–9. [Google Scholar] [CrossRef]
- Sadeer, N.; Rocchetti, G.; Senizza, B.; Montesano, D.; Zengin, G.; Uysal, A.; Jeewon, R.; Lucini, L.; Mahomoodally, M.F. Untargeted Metabolomic Profiling, Multivariate Analysis and Biological Evaluation of the True Mangrove (Rhizophora mucronata Lam.). Antioxidants 2019, 8, 489. [Google Scholar] [CrossRef] [PubMed]
- Bibi Sadeer, N.; Sinan, K.I.; Cziáky, Z.; Jekő, J.; Zengin, G.; Jeewon, R.; Abdallah, H.H.; Rengasamy, K.R.R.; Fawzi Mahomoodally, M. Assessment of the Pharmacological Properties and Phytochemical Profile of Bruguiera gymnorhiza (L.) Lam Using In Vitro Studies, In Silico Docking, and Multivariate Analysis. Biomolecules 2020, 10, 731. [Google Scholar] [CrossRef] [PubMed]
- Manurung, J.; Kappen, J.; Schnitzler, J.; Frolov, A.; Wessjohann, L.A.; Agusta, A.; Muellner-Riehl, A.N.; Franke, K. Analysis of Unusual Sulfated Constituents and Anti-Infective Properties of Two Indonesian Mangroves, Lumnitzera littorea and Lumnitzera racemosa (Combretaceae). Separations 2021, 8, 82. [Google Scholar] [CrossRef]
- Vinh, L.B.; Nguyet, N.T.M.; Yang, S.Y.; Kim, J.H.; Thanh, N.V.; Cuong, N.X.; Nam, N.H.; Minh, C.V.; Hwang, I.; Kim, Y.H. Cytotoxic Triterpene Saponins from the Mangrove Aegiceras corniculatum. Nat. Prod. Res. 2019, 33, 628–634. [Google Scholar] [CrossRef] [PubMed]
- Cruz, S.; Marroquín, M.N.; Cáceres, A. Biological Activity and Chemical Composition of Organic Extracts from Three Guatemalan Mangrove Trees. Int. J. Phytocosmetics Nat. Ingred. 2019, 6, 10. [Google Scholar] [CrossRef]
- Costa, F.D.N.; Jerz, G.; Hewitson, P.; de Figueiredo, F.S.; Ignatova, S. Laguncularia racemosa Phenolics Profiling by Three-Phase Solvent System Step-Gradient Using High-Performance Countercurrent Chromatography with Off-Line Electrospray Mass-Spectrometry Detection. Molecules 2021, 26, 2284. [Google Scholar] [CrossRef]
- Miao, S.; Man, Y.-Q.; Zhou, X.-L.; Yang, L.-J.; Gong, K.-K. Chemical Constituents from Mangrove Plant Sonneratia paracaseolaris. Chin. Tradit. Herb. Drugs 2018, 49, 1025–1030. [Google Scholar] [CrossRef]
- Sasmito, B.; Sulistiyati, T.D.; Hardoko, H. Phytochemicals and Identification of Antioxidant Compounds from Ethanol Extract of Sonneratia alba Leaves and Bark. Russ. J. Agric. Socio-Econ. Sci. 2019, 95, 190–196. [Google Scholar] [CrossRef]
- Li, Z.-H.; Guo, H.; Xu, W.-B.; Ge, J.; Li, X.; Alimu, M.; He, D.-J. Rapid Identification of Flavonoid Constituents Directly from PTP1B Inhibitive Extract of Raspberry (Rubus idaeus L.) Leaves by HPLC–ESI–QTOF–MS-MS. J. Chromatogr. Sci. 2016, 54, 805–810. [Google Scholar] [CrossRef]
- Yi, X.; Jiang, S.; Qin, M.; Liu, K.; Cao, P.; Chen, S.; Deng, J.; Gao, C. Compounds from the Fruits of Mangrove Sonneratia apetala: Isolation, Molecular Docking and Antiaging Effects Using a Caenorhabditis elegans Model. Bioorganic Chem. 2020, 99, 103813. [Google Scholar] [CrossRef]
- Wu, S.-B.; Wen, Y.; Li, X.-W.; Zhao, Y.; Zhao, Z.; Hu, J.-F. Chemical Constituents from the Fruits of Sonneratia caseolaris and Sonneratia ovata (Sonneratiaceae). Biochem. Syst. Ecol. 2009, 37, 1–5. [Google Scholar] [CrossRef]
- Sadhu, S.K.; Ahmed, F.; Ohtsuki, T.; Ishibashi, M. Flavonoids from Sonneratia caseolaris. J. Nat. Med. 2006, 60, 264–265. [Google Scholar] [CrossRef]
- Wu, Y.-L.; Chen, J.-F.; Jiang, L.-Y.; Wu, X.-L.; Liu, Y.-H.; Gao, C.-J.; Wu, Y.; Yi, X.-Q.; Su, Z.-R.; Cai, J.; et al. The Extract of Sonneratia apetala Leaves and Branches Ameliorates Hyperuricemia in Mice by Regulating Renal Uric Acid Transporters and Suppressing the Activation of the JAK/STAT Signaling Pathway. Front. Pharmacol. 2021, 12, 1826. [Google Scholar] [CrossRef]
- Li, Y.; Yu, S.; Liu, D.; Proksch, P.; Lin, W. Inhibitory Effects of Polyphenols toward HCV from the Mangrove Plant Excoecaria agallocha L. Bioorg. Med. Chem. Lett. 2012, 22, 1099–1102. [Google Scholar] [CrossRef] [PubMed]
- De Rosso, M.; Panighel, A.; Vedova, A.; Gardiman, M.; Flamini, R. Characterization of Non-Anthocyanic Flavonoids in Some Hybrid Red Grape Extracts Potentially Interesting for Industrial Uses. Molecules 2015, 20, 18095–18106. [Google Scholar] [CrossRef] [PubMed]
- Saldanha, L.L.; Vilegas, W.; Dokkedal, A.L. Characterization of Flavonoids and Phenolic Acids in Myrcia bella Cambess. Using FIA-ESI-IT-MS(n) and HPLC-PAD-ESI-IT-MS Combined with NMR. Mol. Basel Switz. 2013, 18, 8402–8416. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Bajpai, V.; Kumar, S.; Sharma, K.R.; Kumara, B. Profiling of Gallic and Ellagic Acid Derivatives in Different Plant Parts of Terminalia arjuna by HPLC-ESI-QTOF-MS/MS. Nat. Prod. Commun. 2016, 11, 239–244. [Google Scholar] [CrossRef]
- Khang, D.T.; Anh, L.H.; Thu Ha, P.T.; Tuyen, P.T.; Quan, N.V.; Minh, L.T.; Quan, N.T.; Minh, T.N.; Xuan, T.D.; Khanh, T.D.; et al. Allelopathic Activity of Dehulled Rice and Its Allelochemicals on Weed Germination. Int. Lett. Nat. Sci. 2016, 58, 1–10. [Google Scholar] [CrossRef]
- Zhang, Z.; Cao, J.; Gu, T.; Yang, X.; Peng, Q.; Bai, L.; Li, Y. Co-Planted Barnyardgrass Reduces Rice Yield by Inhibiting Plant above- and Belowground-Growth during Post-Heading Stages. Crop J. 2021, 9, 1198–1207. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, H.; Zhang, J. Root Morphology and Physiology in Relation to the Yield Formation of Rice. J. Integr. Agric. 2012, 11, 920–926. [Google Scholar] [CrossRef]
- Chung, I.-M.; Miller, D.A. Effect of Alfalfa Plant and Soil Extracts on Germination and Growth of Alfalfa. Agron. J. 1995, 87, 762–767. [Google Scholar] [CrossRef]
- Inderjit; Weston, L.A.; Duke, S.O. Challenges, Achievements and Opportunities in Allelopathy Research. J. Plant Interact. 2005, 1, 69–81. [Google Scholar] [CrossRef]
- Xuan, T.D.; Tsuzuki, E.; Hiroyuki, T.; Mitsuhiro, M.; Khanh, T.D.; Chung, I.-M. Evaluation on Phytotoxicity of Neem (Azadirachta indica. A. Juss) to Crops and Weeds. Crop Prot. 2004, 4, 335–345. [Google Scholar] [CrossRef]
- Hasegawa, A.; Oyanagi, T.; Minagawa, R.; Fujii, Y.; Sasamoto, H. An Inverse Relationship between Allelopathic Activity and Salt Tolerance in Suspension Cultures of Three Mangrove Species, Sonneratia alba, S. caseolaris and S. ovata: Development of a Bioassay Method for Allelopathy, the Protoplast Co-Culture Method. J. Plant Res. 2014, 127, 755–761. [Google Scholar] [CrossRef]
- Li, Z.-H.; Wang, Q.; Ruan, X.; Pan, C.-D.; Jiang, D.-A. Phenolics and Plant Allelopathy. Mol. Basel Switz. 2010, 15, 8933–8952. [Google Scholar] [CrossRef]
- Lang, T.; Wei, P.; Chen, X.; Fu, Y.; Tam, N.F.; Hu, Z.; Chen, Z.; Li, F.; Zhou, H. Microcosm Study on Allelopathic Effects of Leaf Litter Leachates and Purified Condensed Tannins from Kandelia obovata on Germination and Growth of Aegiceras corniculatum. Forests 2021, 12, 1000. [Google Scholar] [CrossRef]
- Liu, D.L.; An, M.; Johnson, I.R.; Lovett, J.V. Mathematical Modeling of Allelopathy. III. A Model for Curve-Fitting Allelochemical Dose Responses. Nonlinearity Biol. Toxicol. Med. 2003, 1, 37–50. [Google Scholar] [CrossRef]
- An, M.; Johnson, I.R.; Lovett, J.V. Mathematical Modeling of Allelopathy: Biological Response to Allelochemicals and Its Interpretation. J. Chem. Ecol. 1993, 19, 2379–2388. [Google Scholar] [CrossRef]
- Liu, Y.; Li, F.; Huang, Q. Allelopathic Effects of Gallic Acid from Aegiceras corniculatum on Cyclotella caspia. J. Environ. Sci. 2013, 25, 776–784. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, S.; Wu, J.; Yang, J. Chemical Constituents of Aegiceras corniculatum. Nat. Prod. 2007, 127–133. [Google Scholar]
- Rajeswari, K.; Rao, T.B. Aegiceras corniculatum Linn (Myrsinaceae). J. Chem. Pharm. Res. 2015, 12, 305–316. [Google Scholar]
- Ponnapalli, M.G.; Annam, S.C.V.A.R.; Ravirala, S.; Sukki, S.; Ankireddy, M.; Tuniki, V.R. Unusual Isomeric Corniculatolides from Mangrove, Aegiceras corniculatum. J. Nat. Prod. 2012, 75, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Vinh, L.B.; Phong, N.V.; Ali, I.; Dan, G.; Koh, Y.S.; Anh, H.L.T.; Van Anh, D.T.; Yang, S.Y.; Kim, Y.H. Identification of Potential Anti-Inflammatory and Melanoma Cytotoxic Compounds from Aegiceras corniculatum. Med. Chem. Res. 2020, 29, 2020–2027. [Google Scholar] [CrossRef]
- Reigosa, M.J.; Roger, M.J.R.; Gonzâalez, L.; Pedrol, N.; Gonzalez, L.; González, L. Allelopathy: A Physiological Process with Ecological Implications; Springer Science & Business Media: Berlin, Germany, 2006. [Google Scholar]
- Ghimire, B.K.; Ghimire, B.; Yu, C.Y.; Chung, I.-M. Allelopathic and Autotoxic Effects of Medicago sativa—Derived Allelochemicals. Plants 2019, 8, 233. [Google Scholar] [CrossRef] [PubMed]
- Oleszek, W.; Price, K.R.; Colquhoun, I.J.; Jurzysta, M.; Ploszynski, M.; Fenwick, G.R. Isolation and Identification of Alfalfa (Medicago sativa L.) Root Saponins: Their Activity in Relation to a Fungal Bioassay. J. Agric. Food Chem. 1990, 38, 1810–1817. [Google Scholar] [CrossRef]
- Ladhari, A.; Gaaliche, B.; Zarrelli, A.; Ghannem, M.; Ben Mimoun, M. Allelopathic Potential and Phenolic Allelochemicals Discrepancies in Ficus carica L. Cultivars. South Afr. J. Bot. 2020, 130, 30–44. [Google Scholar] [CrossRef]
- Islam, M.T. Chemical Profile and Biological Activities of Sonneratia apetala (Buch.-Ham.). Adv. Tradit. Med. 2020, 20, 123–132. [Google Scholar] [CrossRef]
- Quraishi, F.M.; Jadhav, B.L. A Review of Phytochemical and Biological Studies on Sonneratia apetala. World J. Adv. Healthc. Res. 2018, 2, 3. [Google Scholar]
- Jaimini, D.; Sarkar, C.; Shabnam, A.A.; Jadhav, B.L. Evaluation of Antibacterial Properties of Mangrove Plant Sonneratia apetala Buch. Ham Leaf. 2011, 4, 1683–1686. [Google Scholar]
- Prabhu Teja, V.; Ravishankar, K. Preliminary Phytochemical Investigation and in Vitro Antimicrobial Activity of Ethanolic Extract of Sonneratia apetala Plant. Int. Res. J. Pharm. 2013, 4, 84–87. [Google Scholar] [CrossRef]
- Patra, J.K.; Das, S.K.; Thatoi, H. Phytochemical Profiling and Bioactivity of a Mangrove Plant, Sonneratia apetala, from Odisha Coast of India. Chin. J. Integr. Med. 2015, 21, 274–285. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, H.; Anwar, T.; Ali, Q.; Haider, M.Z.; Habib, N.; Fatima, S.; Waseem, M.; Bibi, Y.; Arshad, M.; Adkins, S.W. Isolation of Natural Herbicidal Compound from Lantana camara. Int. J. Environ. Anal. Chem. 2021, 101, 631–638. [Google Scholar] [CrossRef]
- Kalinova, J.; Vrchotova, N.; Triska, J. Exudation of Allelopathic Substances in Buckwheat (Fagopyrum esculentum Moench). J. Agric. Food Chem. 2007, 55, 6453–6459. [Google Scholar] [CrossRef] [PubMed]
- Basile, A.; Sorbo, S.; López-Sáez, J.A.; Castaldo Cobianchi, R. Effects of Seven Pure Flavonoids from Mosses on Germination and Growth of Tortula muralis HEDW (Bryophyta) and Raphanus sativus L (Magnoliophyta). Phytochemistry 2003, 62, 1145–1151. [Google Scholar] [CrossRef]
- Correia-da-Silva, M.; Sousa, E.; Pinto, M.M.M. Emerging Sulfated Flavonoids and Other Polyphenols as Drugs: Nature as an Inspiration. Med. Res. Rev. 2014, 34, 223–279. [Google Scholar] [CrossRef]
- Owczarek, A.; Różalski, M.; Krajewska, U.; Olszewska, M.A. Rare Ellagic Acid Sulphate Derivatives from the Rhizome of Geum rivale L.—Structure, Cytotoxicity, and Validated HPLC-PDA Assay. Appl. Sci. 2017, 7, 400. [Google Scholar] [CrossRef]
- Terashima, S.; Shimizu, M.; Nakayama, H.; Ishikura, M.; Ueda, Y.; Imai, K.; Suzui, A.; Morita, N. Studies on Aldose Reductase Inhibitors from Medicinal Plant of “Sinfito,” Potentilla candicans, and Further Synthesis of Their Related Compounds. Chem. Pharm. Bull. 1990, 38, 2733–2736. [Google Scholar] [CrossRef]
- Zhao, D.-K.; Shi, Y.-N.; Petrova, V.; Yue, G.G.L.; Negrin, A.; Wu, S.-B.; D’Armiento, J.M.; Lau, C.B.S.; Kennelly, E.J. Jaboticabin and Related Polyphenols from Jaboticaba (Myrciaria cauliflora) with Anti-Inflammatory Activity for Chronic Obstructive Pulmonary Disease. J. Agric. Food Chem. 2019, 67, 1513–1520. [Google Scholar] [CrossRef]
- Balk, M.; Keuskamp, J.A.; Laanbroek, H.J. Potential for Sulfate Reduction in Mangrove Forest Soils: Comparison between Two Dominant Species of the Americas. Front. Microbiol. 2016, 7, 1855. [Google Scholar] [CrossRef]
- Uddin, S.J.; Jason, T.L.H.; Beattie, K.D.; Grice, I.D.; Tiralongo, E. (2S,3S)-Sulfated Pterosin C, a Cytotoxic Sesquiterpene from the Bangladeshi Mangrove Fern Acrostichum aureum. J. Nat. Prod. 2011, 74, 2010–2013. [Google Scholar] [CrossRef]
- Lee, S.I.; Park, K.W.; Won, O.J.; Park, S.H.; Eom, M.Y.; Hwang, K.S.; Kim, Y.T.; Pyon, J.Y. Effect of rice bran and its mixture with pine leaves on efficacy of weed control and growth and yield of rice in paddy fields. Korean J. Agric. Sci. 2015, 42, 111–116. [Google Scholar] [CrossRef]
- Quang, N.H.; Quinn, C.H.; Stringer, L.C.; Carrie, R.; Hackney, C.R.; Van Hue, L.T.; Van Tan, D.; Nga, P.T.T. Multi-Decadal Changes in Mangrove Extent, Age and Species in the Red River Estuaries of Viet Nam. Remote Sens. 2020, 12, 2289. [Google Scholar] [CrossRef]
- Tomlinson, P.B. The Botany of Mangroves, 2nd ed.; Cambridge University Press: Cambridge, UK, 2016. [Google Scholar]
- Spalding, M.; Kainuma, M.; Collins, L. World Atlas of Mangroves; Earthscan: London, UK, 2010. [Google Scholar]
- Sadeghloo, A.; Asghari, J.; Ghaderi-Far, F. Seed Germination and Seedling Emergence of Velvetleaf (Abutilon theophrasti) and Barnyardgrass (Echinochloa crus-galli). Planta Daninha 2013, 31, 259–266. [Google Scholar] [CrossRef]
- Herranz, J.M.; Ferrandis, P.; Copete, M.A.; Duro, E.M.; Zalacaín, A. Effect of Allelopathic Compounds Produced by Cistus ladanifer on Germination of 20 Mediterranean Taxa. Plant Ecol. 2006, 184, 259–272. [Google Scholar] [CrossRef]
- Vyvyan, J.R. Allelochemicals as Leads for New Herbicides and Agrochemicals. Tetrahedron 2002, 58, 1631–1646. [Google Scholar] [CrossRef]
- Sebastian, J.; Yee, M.-C.; Viana, W.G.; Rellán-Álvarez, R.; Feldman, M.; Priest, H.D.; Trontin, C.; Lee, T.; Jiang, H.; Baxter, I.; et al. Grasses Suppress Shoot-Borne Roots to Conserve Water during Drought. Proc. Natl. Acad. Sci. USA 2016, 113, 8861–8866. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Michel, T.; Khlif, I.; Kanakis, P.; Termentzi, A.; Allouche, N.; Halabalaki, M.; Skaltsounis, A.-L. UHPLC-DAD-FLD and UHPLC-HRMS/MS Based Metabolic Profiling and Characterization of Different Olea europaea Organs of Koroneiki and Chetoui Varieties. Phytochem. Lett. 2015, 11, 424–439. [Google Scholar] [CrossRef]
- Brakni, R.; Ali Ahmed, M.; Burger, P.; Schwing, A.; Michel, G.; Pomares, C.; Hasseine, L.; Boyer, L.; Fernandez, X.; Landreau, A.; et al. UHPLC-HRMS/MS Based Profiling of Algerian Lichens and Their Antimicrobial Activities. Chem. Biodivers. 2018, 15, e1800031. [Google Scholar] [CrossRef]


| RAE on Germination Rate (%) | RAE on Root Length (%) | |||||
|---|---|---|---|---|---|---|
| Df | F-Value | p-Value | Df | F-Value | p-Value | |
| Echinochloa crus-galli | ||||||
| Source species (S) | 7 | 13.3 | *** | 7 | 201.8 | *** |
| Dose (D) | 1 | 6.1 | * | 1 | 173.8 | *** |
| S × D | 7 | 1.2 | 7 | 11.0 | *** | |
| Residuals | 64 | 892 | ||||
| Oryza sativa | ||||||
| Source species (S) | 7 | 137.1 | *** | 7 | 287.6 | *** |
| Dose (D) | 1 | 13.3 | *** | 1 | 301.6 | *** |
| S × D | 7 | 4.7 | *** | 7 | 31.6 | *** |
| Residuals | 64 | 1285 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dhaou, D.; Baldy, V.; Van Tan, D.; Malachin, J.-R.; Pouchard, N.; Roux, A.; Dupouyet, S.; Greff, S.; Culioli, G.; Michel, T.; et al. Allelopathic Potential of Mangroves from the Red River Estuary against the Rice Weed Echinochloa crus-galli and Variation in Their Leaf Metabolome. Plants 2022, 11, 2464. https://doi.org/10.3390/plants11192464
Dhaou D, Baldy V, Van Tan D, Malachin J-R, Pouchard N, Roux A, Dupouyet S, Greff S, Culioli G, Michel T, et al. Allelopathic Potential of Mangroves from the Red River Estuary against the Rice Weed Echinochloa crus-galli and Variation in Their Leaf Metabolome. Plants. 2022; 11(19):2464. https://doi.org/10.3390/plants11192464
Chicago/Turabian StyleDhaou, Dounia, Virginie Baldy, Dao Van Tan, Jean-Rémi Malachin, Nicolas Pouchard, Anaïs Roux, Sylvie Dupouyet, Stéphane Greff, Gérald Culioli, Thomas Michel, and et al. 2022. "Allelopathic Potential of Mangroves from the Red River Estuary against the Rice Weed Echinochloa crus-galli and Variation in Their Leaf Metabolome" Plants 11, no. 19: 2464. https://doi.org/10.3390/plants11192464
APA StyleDhaou, D., Baldy, V., Van Tan, D., Malachin, J. -R., Pouchard, N., Roux, A., Dupouyet, S., Greff, S., Culioli, G., Michel, T., Fernandez, C., & Bousquet-Mélou, A. (2022). Allelopathic Potential of Mangroves from the Red River Estuary against the Rice Weed Echinochloa crus-galli and Variation in Their Leaf Metabolome. Plants, 11(19), 2464. https://doi.org/10.3390/plants11192464