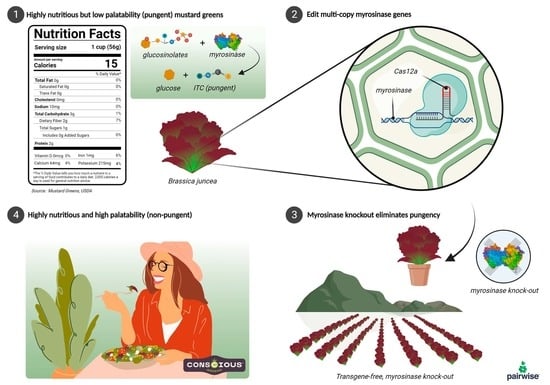
Targeted Mutagenesis of the Multicopy Myrosinase Gene Family in Allotetraploid Brassica juncea Reduces Pungency in Fresh Leaves across Environments
Abstract
:1. Introduction
2. Results
2.1. Bioinformatics Analysis
2.2. Edit Characterization and Advancement of Edited Lines
2.3. Structural Modeling of Edited B05 Myrosinase
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Genome Assembly of B. juncea cultivar Red Giant
5.2. RNAseq Analysis
5.3. Myrosinase Gene Bioinformatics
5.4. Vector Design
5.5. Plant Transformation
5.6. Controlled Environments and Field Growth Conditions
5.7. Deep Sequencing of Edited Plants
5.8. Probe Design for Myrosinase Gene Characterization and Transgene Presence/Absence Using Target-Capture Sequencing
5.9. Target-Capture Library Preparation and Sequencing
5.10. Bioinformatics Procedure of Plasmid Target-Capture Sequencing for Plasmid Purity Test
5.11. Bioinformatics Procedure of Gene Target-Capture Sequencing for Edit Characterization
5.12. Phenotypic Evaluation
5.13. GRA Assay
5.14. Sensory Assay for Evaluating Pungency
5.15. Structural Modeling of Unedited and Edited Myrosinase Encoded on Chromosome B05
5.16. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Keatinge, J.D.H.; Yang, R.-Y.; Hughes, J.D.A.; Easdown, W.J.; Holmer, R. The Importance of Vegetables in Ensuring Both Food and Nutritional Security in Attainment of the Millennium Development Goals. Food Secur. 2011, 3, 491–501. [Google Scholar] [CrossRef]
- Kumar, D.; Kumar, S.; Shekhar, C. Nutritional Components in Green Leafy Vegetables: A Review. JPP 2020, 5, 2498–2502. [Google Scholar]
- Lee, S.H.; Moore, L.V.; Park, S.; Harris, D.M.; Blanck, H.M. Adults Meeting Fruit and Vegetable Intake Recommendations—United States, 2019. Morb. Mortal. Wkly. Rep. 2022, 71, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.J.; Basset, G.; Sandoya, G. Nutritional Benefits of Lettuce Consumed at Recommended Portion Sizes. Edis 2021, HS1416, 1–8. [Google Scholar] [CrossRef]
- Kulkarni, K.D. Food, Culture, and Diabetes in the United States. Clin. Diabetes 2004, 22, 190–192. [Google Scholar] [CrossRef]
- Natesh, H.N.; Abbey, L.; Asiedu, S.K. An Overview of Nutritional and Anti Nutritional Factors in Green Leafy Vegetables. Hortic. Int. J. 2017, 1, 00011. [Google Scholar] [CrossRef]
- Micozzi, M.S.; Beecher, G.R.; Taylor, P.R.; Khachik, F. Carotenoid Analyses of Selected Raw and Cooked Foods Associated With a Lower Risk for Cancer. JNCI J. Natl. Cancer Inst. 1990, 82, 282–285. [Google Scholar] [CrossRef]
- Noia, J.D. Defining Powerhouse Fruits and Vegetables: A Nutrient Density Approach. Prev. Chronic. Dis. 2014, 11, E95. [Google Scholar] [CrossRef]
- Zhang, C.; Wei, Y.; Xiao, D.; Gao, L.; Lyu, S.; Hou, X.; Bouuema, G. Transcriptomic and Proteomic Analyses Provide New Insights into the Regulation Mechanism of Low-Temperature-Induced Leafy Head Formation in Chinese Cabbage. J. Proteomics 2016, 144, 1–10. [Google Scholar] [CrossRef]
- Gu, A.; Meng, C.; Chen, Y.; Wei, L.; Dong, H.; Lu, Y.; Wang, Y.; Chen, X.; Zhao, J.; Shen, S. Coupling Seq-BSA and RNA-Seq Analyses Reveal the Molecular Pathway and Genes Associated with Heading Type in Chinese Cabbage. Front. Genet. 2017, 8, 176. [Google Scholar] [CrossRef]
- Liang, J.; Liu, B.; Wu, J.; Cheng, F.; Wang, X. Genetic Variation and Divergence of Genes Involved in Leaf Adaxial-Abaxial Polarity Establishment in Brassica Rapa. Front. Plant Sci. 2016, 7, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fenwick, G.R.; Heaney, R.K.; Mullin, W.J.; VanEtten, C.H. Glucosinolates and Their Breakdown Products in Food and Food Plants. CRC Crit. Rev. Food Sci. Nutr. 1983, 18, 123–201. [Google Scholar] [CrossRef] [PubMed]
- Lüthy, B.; Matile, P. The Mustard Oil Bomb: Rectified Analysis of the Subcellular Organisation of the Myrosinase System. Biochem. Physiol. Pflanz. 1984, 179, 5–12. [Google Scholar] [CrossRef]
- Graham, N.; Patil, G.; Bubeck, D.M.; Dobert, R.C.; Glenn, K.C.; Gutsche, A.T.; Kumar, S.; Lindbo, J.A.; Maas, L.; May, G.D.; et al. Plant Genome Editing and the Relevance of Off-Target Changes. Plant Physiol. 2020, 183, 1453–1471. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Fräbel, S.; Lawit, S.J.; Nie, J.; Schwark, D.G.; Poorten, T.J.; Graham, N.D. Progress in precise and predictable genome editing in plants with base editing. In Genome Editing for Precision Crop Breeding, 1st ed.; Willmann, M.R., Ed.; Burleigh Dodds Science Publishing: London, UK, 2021; Volume 1, pp. 123–146. [Google Scholar] [CrossRef]
- Rees, H.A.; Liu, D.R. Base Editing: Precision Chemistry on the Genome and Transcriptome of Living Cells. Nat. Rev. Genet. 2018, 19, 770–788. [Google Scholar] [CrossRef]
- Lin, Q.; Zong, Y.; Xue, C.; Wang, S.; Jin, S.; Zhu, Z.; Wang, Y.; Anzalone, A.V.; Raguram, A.; Doman, J.L.; et al. Prime Genome Editing in Rice and Wheat. Nat. Biotechnol. 2020, 38, 582–585. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-Replace Genome Editing without Double-Strand Breaks or Donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Bell, L.; Oloyede, O.O.; Lignou, S.; Wagstaff, C.; Methven, L. Taste and Flavor Perceptions of Glucosinolates, Isothiocyanates, and Related Compounds. Mol. Nutr. Food Res. 2018, 62, 1700990. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [Green Version]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Kumar, S.; Sangwan, S.; Yadav, I.S.; Yadav, R. Protein Modeling and Active Site Binding Mode Interactions of Myrosinase–Sinigrin in Brassica Juncea—An in Silico Approach. J. Mol. Graph. Model. 2011, 29, 740–746. [Google Scholar] [CrossRef] [PubMed]
- Dosz, E.B.; Ku, K.-M.; Juvik, J.A.; Jeffery, E.H. Total Myrosinase Activity Estimates in Brassica Vegetable Produce. J. Agric. Food Chem. 2014, 62, 8094–8100. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Lawit, S.J.; Weers, B.; Sun, J.; Mongar, N.; Hemert, J.V.; Melo, R.; Meng, X.; Rupe, M.; Clapp, J.; et al. Overexpression of Zmm28 Increases Maize Grain Yield in the Field. Proc. Natl. Acad. Sci. USA 2019, 116, 23850–23858. [Google Scholar] [CrossRef] [PubMed]
- Zastrow-Hayes, G.M.; Lin, H.; Sigmund, A.L.; Hoffman, J.L.; Alarcon, C.M.; Hayes, K.R.; Richmond, T.A.; Jeddeloh, J.A.; May, G.D.; Beatty, M.K. Southern-by-Sequencing: A Robust Screening Approach for Molecular Characterization of Genetically Modified Crops. Plant Genome 2015, 8, eplantgenome2014.08.0037. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; DiMaio, F.; Wang, R.Y.-R.; Kim, D.; Miles, C.; Brunette, T.; Thompson, J.; Baker, D. High-Resolution Comparative Modeling with RosettaCM. Structure 2013, 21, 1735–1742. [Google Scholar] [CrossRef]
- Conway, P.; Tyka, M.D.; DiMaio, F.; Konerding, D.E.; Baker, D. Relaxation of Backbone Bond Geometry Improves Protein Energy Landscape Modeling. Protein Sci. 2014, 23, 47–55. [Google Scholar] [CrossRef]
- Jones, D.T. Protein Secondary Structure Prediction Based on Position-Specific Scoring Matrices1 1Edited by G. Von Heijne. J. Mol. Biol. 1999, 292, 195–202. [Google Scholar] [CrossRef]
- Berka, K.; Sehnal, D.; Bazgier, V.; Pravda, L.; Svobodova-Varekova, R.; Otyepka, M.; Koca, J. Mole 2.5—Tool for Detection and Analysis of Macromolecular Pores and Channels. Biophys. J. 2017, 112, 292a–293a. [Google Scholar] [CrossRef]
- Rask, L.; Andréasson, E.; Ekbom, B.; Eriksson, S.; Pontoppidan, B.; Meijer, J. Myrosinase: Gene Family Evolution and Herbivore Defense in Brassicaceae. Plant Mol. Biol. 2000, 42, 93–114. [Google Scholar] [CrossRef]
- Barth, C.; Jander, G. Arabidopsis Myrosinases TGG1 and TGG2 Have Redundant Function in Glucosinolate Breakdown and Insect Defense. Plant J. 2006, 46, 549–562. [Google Scholar] [CrossRef] [PubMed]
- Burmeister, W.P.; Cottaz, S.; Driguez, H.; Iori, R.; Palmieri, S.; Henrissat, B. The Crystal Structures of Sinapis Alba Myrosinase and a Covalent Glycosyl–Enzyme Intermediate Provide Insights into the Substrate Recognition and Active-Site Machinery of an S-Glycosidase. Structure 1997, 5, 663–676. [Google Scholar] [CrossRef]
- Haun, W.; Coffman, A.; Clasen, B.M.; Demorest, Z.L.; Lowy, A.; Ray, E.; Retterath, A.; Stoddard, T.; Juillerat, A.; Cedrone, F.; et al. Improved Soybean Oil Quality by Targeted Mutagenesis of the Fatty Acid Desaturase 2 Gene Family. Plant Biotechnol. J. 2014, 12, 934–940. [Google Scholar] [CrossRef] [PubMed]
- Pham, A.-T.; Lee, J.-D.; Shannon, J.G.; Bilyeu, K.D. A Novel FAD2-1 A Allele in a Soybean Plant Introduction Offers an Alternate Means to Produce Soybean Seed Oil with 85% Oleic Acid Content. Theor. Appl. Genet. 2011, 123, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Concepcion, G.T.; Feng, X.; Zhang, H.; Li, H. Haplotype-Resolved de Novo Assembly Using Phased Assembly Graphs with Hifiasm. Nat. Methods 2021, 18, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, Scalable Generation of High-quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Nagy, E.D.; Stevens, J.L.; Yu, N.; Hubmeier, C.S.; LaFaver, N.; Gillespie, M.; Gardunia, B.; Cheng, Q.; Johnson, S.; Vaughn, A.L.; et al. Novel Disease Resistance Gene Paralogs Created by CRISPR/Cas9 in Soy. Plant Cell Rep. 2021, 40, 1047–1058. [Google Scholar] [CrossRef]
- Jiang, W.; Zhou, H.; Bi, H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/SgRNA-Mediated Targeted Gene Modification in Arabidopsis, Tobacco, Sorghum and Rice. Nucleic Acids Res. 2013, 41, e188. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Juarez, B. Re: Confirmation of the Regulatory Status of Genome Edited Brassica Juncea Lines with Improved Flavor. Available online: https://www.aphis.usda.gov/biotechnology/downloads/reg_loi/20-168-07_air_response_signed.pdf (accessed on 20 August 2020).
- Yang, J.; Liu, D.; Wang, X.; Ji, C.; Cheng, F.; Liu, B.; Hu, Z.; Chen, S.; Pental, D.; Ju, Y.; et al. The Genome Sequence of Allopolyploid Brassica Juncea and Analysis of Differential Homoeolog Gene Expression Influencing Selection. Nat. Genet. 2016, 48, 1225–1232. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows–Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Garrison, E.; Marth, G. Haplotype-Based Variant Detection from Short-Read Sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- Layer, R.M.; Chiang, C.; Quinlan, A.R.; Hall, I.M. LUMPY: A Probabilistic Framework for Structural Variant Discovery. Genome Biol. 2014, 15, R84. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative Genomics Viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Remmert, M.; Biegert, A.; Hauser, A.; Söding, J. HHblits: Lightning-Fast Iterative Protein Sequence Searching by HMM-HMM Alignment. Nat. Methods 2012, 9, 173. [Google Scholar] [CrossRef]
- Söding, J. Protein Homology Detection by HMM–HMM Comparison. Bioinformatics 2005, 21, 951–960. [Google Scholar] [CrossRef]
- Gront, D.; Kulp, D.W.; Vernon, R.M.; Strauss, C.E.M.; Baker, D. Generalized Fragment Picking in Rosetta: Design, Protocols and Applications. PLoS ONE 2011, 6, e23294. [Google Scholar] [CrossRef]
- Hosseinzadeh, P.; Bhardwaj, G.; Mulligan, V.K.; Shortridge, M.D.; Craven, T.W.; Pardo-Avila, F.; Rettie, S.A.; Kim, D.E.; Silva, D.-A.; Ibrahim, Y.M.; et al. Comprehensive Computational Design of Ordered Peptide Macrocycles. Science 2017, 358, 1461–1466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shapiro, S.S.; Wilk, M.B. An Analysis of Variance Test for Normality (Complete Samples). Biometrika 1965, 52, 591–611. [Google Scholar] [CrossRef]
- Kolmogorov–Smirnov Test. In The Concise Encyclopedia of Statistics; Springer: New York, NY, USA, 2008; p. 344. ISBN 9780387317427.
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Available online: http://www.R-project.org/ (accessed on 10 October 2020).
- RStudio Team. RStudio: Integrated Development for R. RStudio. Available online: http://www.rstudio.com/ (accessed on 1 February 2022).
- Wickham, H. Ggplot2 Use R! 1st ed.; Springer: Cham, Switzerland, 2016; pp. 147–168. [Google Scholar] [CrossRef]
- Russel, V. Emmeans: Estimated Marginal Means, Aka Least-Squares Means. R Package Version 1.7.2. Available online: https://CRAN.R-project.org/package=emmeans (accessed on 27 March 2022).
- Dawson, C. Ggprism: A “ggplot2” Extension Inspired by “GraphPad Prism”. R Package Version 1.0.3. Available online: https://CRAN.R-project.org/package=ggprism (accessed on 3 May 2022).
- Goode, K.; Rey, K. GgResidpanel: Panels and Interactive Versions of Diagnostic Plots Using “Ggplot2”. R Package Version 0.3.0.9000. Available online: https://goodekat.github.io/ggResidpanel/ (accessed on 31 May 2019).








| Gene | Edit | Putative Edit Effect |
|---|---|---|
| A01.1_rg | 7 bp deletion | Pre-mature stop (at exon 6) |
| A02.1_rg | 365 bp deletion | Loss of at least 2 active site residues; pre-mature stop (at exon 7 exons 5 and 6 partially deleted) |
| A02.2_rg | 1923 bp deletion | Loss of multiple exons upstream of the spacer (exons 1–5 deleted, exon 6 partially deleted) |
| A02.3_rg | 10 bp deletion, 3971 bp deletion | Loss of multiple exons downstream of the spacer (exon 6 partially deleted, exons 7–12 deleted) |
| A02.4_rg | 93 bp deletion, 8 bp deletion | Pre-mature stop (at exon 6) |
| A03.1_rg | 7 bp deletion, 7 bp deletion | Pre-mature stop (at exon 6) |
| A09.1_rg | 8 bp deletion, 13 bp deletion, 4 bp deletion | Pre-mature stop (at exon 7) |
| A09_cl2.1_rg | 8 bp deletion | Pre-mature stop (at exon 6) |
| A09_cl2.2_rg | 8 bp deletion, 6 bp deletion | Pre-mature stop (at exon 6) |
| B04_cl1.1_rg | whole gene deletion | Whole gene deletion |
| B04_cl1.2_rg | whole gene deletion | Whole gene deletion |
| B04_cl1.3_rg | B04 gene chimera via inversion | Pre-mature stop (at exon 6) |
| B04_cl1.4_rg | B04 gene chimera via inversion | Pre-mature stop (at exon 6) |
| B04_cl1.5_rg | B04 gene chimera via inversion | Pre-mature stop (at exon 6) |
| B04_cl2.1_rg | 316 bp deletion | Loss of 1 active site residue |
| B07.1_rg | 8 bp deletion, 5 bp deletion | Pre-mature stop (at exon 7) |
| B05 | 21 bp deletion | 7 aa deletion including one active site residue (Q>K in exon 6) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karlson, D.; Mojica, J.P.; Poorten, T.J.; Lawit, S.J.; Jali, S.; Chauhan, R.D.; Pham, G.M.; Marri, P.; Guffy, S.L.; Fear, J.M.; et al. Targeted Mutagenesis of the Multicopy Myrosinase Gene Family in Allotetraploid Brassica juncea Reduces Pungency in Fresh Leaves across Environments. Plants 2022, 11, 2494. https://doi.org/10.3390/plants11192494
Karlson D, Mojica JP, Poorten TJ, Lawit SJ, Jali S, Chauhan RD, Pham GM, Marri P, Guffy SL, Fear JM, et al. Targeted Mutagenesis of the Multicopy Myrosinase Gene Family in Allotetraploid Brassica juncea Reduces Pungency in Fresh Leaves across Environments. Plants. 2022; 11(19):2494. https://doi.org/10.3390/plants11192494
Chicago/Turabian StyleKarlson, Dale, Julius P. Mojica, Thomas J. Poorten, Shai J. Lawit, Sathya Jali, Raj Deepika Chauhan, Gina M. Pham, Pradeep Marri, Sharon L. Guffy, Justin M. Fear, and et al. 2022. "Targeted Mutagenesis of the Multicopy Myrosinase Gene Family in Allotetraploid Brassica juncea Reduces Pungency in Fresh Leaves across Environments" Plants 11, no. 19: 2494. https://doi.org/10.3390/plants11192494
APA StyleKarlson, D., Mojica, J. P., Poorten, T. J., Lawit, S. J., Jali, S., Chauhan, R. D., Pham, G. M., Marri, P., Guffy, S. L., Fear, J. M., Ochsenfeld, C. A., Chapman, T. A., Casamali, B., Venegas, J. P., Kim, H. J., Call, A., Sublett, W. L., Mathew, L. G., Shariff, A., ... Rapp, R. (2022). Targeted Mutagenesis of the Multicopy Myrosinase Gene Family in Allotetraploid Brassica juncea Reduces Pungency in Fresh Leaves across Environments. Plants, 11(19), 2494. https://doi.org/10.3390/plants11192494