Physiological and Molecular Responses of Pyrus pyraster Seedlings to Salt Treatment Analyzed by miRNA and Cytochrome P450 Gene-Based Markers
Abstract
:1. Introduction
2. Material and Methods
2.1. Plant Material
2.2. Experimental Design
- Mn = moisture content (%) of the material n;
- Ww = wet weight of the sample;
- Wd = weight of the sample after drying.
2.3. Measurement and Analysis of Plant Parameters
2.4. Leaf Gas Exchange
2.5. Leaf Water Potential and Relative Water Content
- FW = fresh weight;
- DW = dry weight;
- SW = weight after full saturation of the leaf samples.
2.6. Statistical Analysis
2.7. MicroRNA-Based Assay
2.8. PBA-Based Assay
2.9. PBA Expression Analysis
3. Results
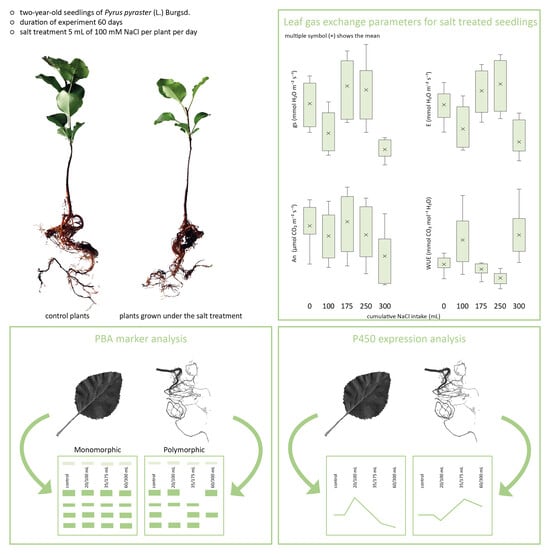
3.1. Plant Growth and Mass Accumulation under Salt Treatment
3.2. Leaf Water Status under Salt Treatment
3.3. Effect of Salinity on Leaf Gas Exchange
3.4. Effect of Salinity on Plant Genome Response, Analyzed by DNA-Based Markers and Cytochrome P450 Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Stephan, B.R.; Wagner, I.; Kleinschmit, J. EUFORGEN Technical Guidelines for Genetic Conservation and Use for Wild Apple and Pear (Malus sylvestris and Pyrus pyraster); International Plant Genetic Resources Institute: Rome, Italy, 2003. [Google Scholar]
- Wagner, I.; Büttner, R. Hybridization in wild pear (Pyrus pyraster) from various regions in Germany and from Luxembourg with respect to Pyrus × communis. In Proceedings of the III International Symposium on Horticulture in Europe-SHE2016, Chania, Greece, 17–21 October 2016; pp. 427–434. [Google Scholar]
- Ellenberg, H. Vegetation Mitteleuropas mit den Alpen in Ökologischer Sicht, 3rd ed.; Ulmer Verlag: Stuttgart, Germany, 1983. [Google Scholar]
- Peniašteková, M. Pyrus L. Hruška. In Flóra Slovenska IV/3; Bertová, L., Ed.; Veda: Bratislava, Slovakia, 1992; pp. 381–388. [Google Scholar]
- Hegi, G. Illustrierte Flora von Mitteleuropa; Blackwell Wissenschaft-Verlag: Berlin, Germany, 1995; pp. 284–286. [Google Scholar]
- Paganová, V. Wild pear Pyrus pyraster (L.) Burgsd. requirements on environmental conditions. Ekológia 2003, 22, 225–241. [Google Scholar]
- Milner, E. Trees of Britain and Ireland; Natural History Museum: London, UK, 2011. [Google Scholar]
- Kleinschmit, J.; Stephan, R.; Wagner, I. Conservation of genetic resources of wild fruit species (Prunus avium, Malus sylvestris and Pyrus pyraster). In Noble Hardwoods Network Report of the Second Meeting; Turok, J., Collin, E., Demesure, B., Erikson, G., Kleinschmit, J., Rusanen, M., Stephan, R., Eds.; International Plant Genetic Resources Institute: Rome, Italy, 1998; pp. 51–57. [Google Scholar]
- Wolko, Ł.; Bocianowski, J.; Antkowiak, W.; Słomski, R. Genetic diversity and population structure of wild pear (Pyrus pyraster (L.) Burgsd.) in Poland. Open Life Sci. 2014, 10, 19–29. [Google Scholar] [CrossRef]
- Vidaković, A.; Šatović, Z.; Tumpa, K.; Idžojtić, M.; Liber, Z.; Pintar, V.; Radunić, M.; Runjić, T.N.; Runjić, M.; Rošin, J.; et al. Phenotypic Variation in European Wild Pear (Pyrus pyraster (L.) Burgsd.) Populations in the North-Western Part of the Balkan Peninsula. Plants 2022, 11, 335. [Google Scholar] [CrossRef] [PubMed]
- Paganová, V.; Hus, M.; Jureková, Z. Physiological Performance of Pyrus pyraster L. (Burgsd.) and Sorbus torminalis (L.) Crantz Seedlings under Drought Treatment. Plants 2020, 9, 1496. [Google Scholar] [CrossRef]
- Paganová, V.; Hus, M.; Lichtnerová, H. Effect of substrate salinity on growth of juvenile plants Pyrus pyraster (L.) Burgsd. In Plants in Urban Areas and Landscape; Slovak Universty of Agriculture in Nitra: Nitra, Slovakia, 2020; pp. 66–73. [Google Scholar] [CrossRef]
- Paganová, V.; Hus, M.; Lichtnerová, H. Effect of Salt Treatment on the Growth, Water Status, and Gas Exchange of Pyrus pyraster L. (Burgsd.) and Tilia cordata Mill. Seedlings. Horticulturae 2022, 8, 519. [Google Scholar] [CrossRef]
- Ganie, S.A.; Mondal, T.K. Genome-wide development of novel miRNA-based microsatellite markers of rice (Oryza sativa) for genotyping applications. Mol. Breed. 2015, 35, 51. [Google Scholar] [CrossRef]
- Gupta, P.K.; Kulwal, P.L.; Jaiswal, V. Association mapping in crop plants: Opportunities and challenges. Adv. Genet. 2014, 85, 109–147. [Google Scholar] [PubMed]
- Balážová, Ž.; Gálová, Z.; Vivodík, M.; Petrovičová, L.; Hornyák Gregáňová, R. Molecular variability of oat based on genetic specific markers. Potravin. Slovak J. Food Sci. 2017, 11, 332–337. [Google Scholar]
- Inui, H.; Komada, T.; Ohkawa, Y.; Ohkawa, H. Herbicide metabolism and cross-tolerance in transgenic potato plants co-expressing human CYP1A1, CYP2B6 and CYP2C19. Pestic. Biochem. Physiol. 2000, 66, 116–129. [Google Scholar] [CrossRef]
- Xu, Y.; Li, H.; Li, X.; Lin, J.; Wang, Z.; Yang, Q.; Chang, Y. Systematic selection, and validation of appropriate reference genes for gene expression studies by quantitative real-time PCR in pear. Acta Physiol. Plant. 2015, 37, 40. [Google Scholar] [CrossRef]
- Ohkawa, H.; Imaishi, H.; Shiota, N.; Yamada, T.; Inui, H.; Ohkawa, Y. Molecular mechanisms of herbicide resistance with special emphasis on cytochrome P450 monooxygenases. Plant Biotechnol. 1998, 15, 173–176. [Google Scholar] [CrossRef]
- Li, P.; Yang, X.; Wang, H.; Pan, T.; Wang, Y.; Xu, Y.; Xu, C.; Yang, Z. Genetic control of root plasticity in response to salt stress in maize. Theor. Appl. Genet. 2021, 134, 1475–1492. [Google Scholar] [CrossRef]
- Yu, Y.; Wu, G.; Yuan, H.; Cheng, L.; Zhao, D.; Huang, W.; Zhang, S.; Zhang, L.; Chen, H.; Zhang, J.; et al. Identification and characterization of miRNAs and targets in flax (Linum usitatissimum) under saline, alkaline, and saline-alkaline stresses. BMC Plant Biol. 2016, 16, 124. [Google Scholar] [CrossRef]
- Dong, Q.; Hu, B.; Zhang, C. MicroRNAs and Their Roles in Plant Development. Front. Plant Sci. 2022, 13, 824240. [Google Scholar] [CrossRef]
- Xu, X.; Chen, X.; Chen, Y.; Zhang, Q.; Su, L.; Chen, X.; Chen, Y.; Zhang, Z.; Lin, Y.; Lai, Z. Genome-wide identification of miRNAs and their targets during early somatic embryogenesis in Dimocarpus longan Lour. Sci. Rep. 2020, 10, 4626. [Google Scholar] [CrossRef]
- Arazi, T.; Khedia, J. Tomato MicroRNAs and Their Functions. Int. J. Mol. Sci. 2022, 23, 11979. [Google Scholar] [CrossRef]
- Melnikova, N.V.; Dmitriev, A.A.; Belenikin, M.S.; Speranskava, A.S.; Krinitsina, A.A.; Rachinskaia, O.A.; Lakunina, V.A.; Krasnov, G.S.; Snezhkina, A.V.; Sadriidinova, A.F.; et al. Excess fertilizer responsive miRNAs revealed in Linum usitatissimum L. Biochemie 2015, 109, 36–41. [Google Scholar] [CrossRef]
- Fu, D.; Ma, B.; Mason, A.S.; Xiao, M.; Wei, L.; An, Z. MicroRNA-based molecular markers: A novel PCR-based genotyping technique in Brassica species. Plant Breed. 2013, 132, 375–381. [Google Scholar] [CrossRef]
- Yadav, C.B.Y.; Muthamilarasan, M.; Pandey, G.; Prasad, M. Development of novel microRNA-based genetic markers in foxtail millet for genotyping applications in related grass species. Mol. Breed. 2014, 34, 2219–2224. [Google Scholar] [CrossRef]
- Mondal, T.K.; Ganie, S.A. Identification and characterization of salt responsive miRNA-SSR markers in rice (Oryza sativa). Gene 2013, 535, 204–209. [Google Scholar] [CrossRef]
- Niu, Q.; Qian, M.; Liu, G.; Yang, F.; Teng, Y. A genome-wide identification and characterization of mircoRNAs and their targets in ‘Suli’ pear (Pyrus pyrifolia white pear group). Planta 2013, 238, 1095–1112. [Google Scholar] [CrossRef] [PubMed]
- Qian, M.; Ni, J.; Niu, Q.; Bai, S.; Bao, L.; Li, J.; Sun, Y.; Zhang, D.; Teng, Y. Response of miR156-SPL Module during the Red Peel Coloration of Bagging-Treated Chinese Sand Pear (Pyrus pyrifolia Nakai). Front. Physiol. 2017, 8, 550. [Google Scholar] [CrossRef]
- Ma, L.; Li Ch Niu, J.X.; Pei, M.S.; Cao, F.; Quan, S. Cloning and identification of novel miRNAs in the flower organs of Korla fragrant pear at anthesis. J. Hortic. Sci. Biotechnol. 2018, 94, 305–316. [Google Scholar] [CrossRef]
- Wu, J.; Wang, D.; Liu, Y.; Wang, L.; Qiao, X.; Zhang, S. Identification of miRNAs involved in pear fruit development and quality. BMC Genom. 2014, 15, 953. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, X.; Zhang, F.; Hong, N.; Wang, G.; Wang, A.; Wang, L. Identification and characterization of microRNAs from in vitro-grown pear shoots infected with Apple stem grooving virus in response to high temperature using small RNA sequencing. BMC Genom. 2015, 16, 945. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, J.; Liang, Q.; Zhang, Y.; Kang, K.; Wang, W.; Feng, Y.; Wu, S.; Yang, C.; Li, Y. Genome-wide analysis of long noncoding RNAs affecting floral bud dormancy in pears in response to cold stress. Tree Physiol. 2021, 41, 771–790. [Google Scholar] [CrossRef]
- Lapin, M.; Faško, P.; Melo, M.; Štastný, P.; Tomlain, J. Klimatické oblasti. In Atlas Krajiny Slovenskej Republiky; Ministerstvo Životného Prostredia: Banská Bystrica, Slovakia, 2002; p. 344. [Google Scholar]
- Trautmann, N.; Richard, T. Moisture Content. Cornell Waste Management Institute. 1996. Available online: http://compost.css.cornell.edu/calc/moisture_content.html (accessed on 15 February 2019).
- Ostonen, I.; Püttstep, U.; Biel, C.; Alberton, O.; Bakker, M.R.; Löhmus, H.; Majdi, D.; Metcalfe, D.; Olsthoorn, A.F.M.; Pronk, A.; et al. Specific root length as an indicator of environmental changes. Plant Biosyst. 2007, 141, 3426–3442. [Google Scholar] [CrossRef]
- Poorter, H.; Niklas, K.J.; Reich, P.B.; Oleksyn, J.; Poot, P.; Mommer, L. Biomass allocation to leaves, stems and roots: Meta-analyses of interspecific variation and environmental control. New Phytol. 2012, 193, 30–50. [Google Scholar] [CrossRef]
- Parsons, R.; Weyers, J.D.B.; Lawson, T.; Godber, I.M. Rapid and straightforward estimates of photosynthetic characteristics using a portable gas exchange system. Photosynthetica 1998, 34, 265–279. [Google Scholar] [CrossRef]
- Hunt, S. Measurements of photosynthesis and respiration in plants. Physiol. Plant 2003, 117, 314–325. [Google Scholar] [CrossRef]
- Barrs, H.D.; Weatherley, P.E. A re-examination of the relative turgidity technique for estimating water deficit in leaves. Aust. J. Biol. Sci. 1962, 15, 413–428. [Google Scholar] [CrossRef]
- Ražná, K.; Žiarovská, J.; Gálová, Z. MicroRNA-based markers in plant genome response to abiotic stress and their application in plant genotyping. In Non-Coding RNAs, 1st ed.; Intech Open: London, UK, 2020; pp. 71–81. ISBN 978-1-78985-655-2. [Google Scholar]
- Yamanaka, S.; Suzuki, E.; Tanaka, M.; Takeda, Y.; Watanabe, J.A.; Watanabe, K.N. Assessment of cytochrome P450 sequences offers a useful tool for determining genetic diversity in higher plant species. Theor. Appl. Genet. 2003, 108, 1–9. [Google Scholar] [CrossRef]
- Liu, Z.; Cheng, K.; Qin, Z.; Wu, T.; Li, X.; Tu, J.; Yang, F.; Zhu, H.; Yang, L. Selection and validation of suitable reference genes for qRT-PCR analysis in pear leaf tissues under distinct training systems. PLoS ONE 2018, 13, e0202472. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Kozlowski, T.T. Responses of woody plants to flooding and salinity. Tree Physiol. 1997, 17, 490. [Google Scholar] [CrossRef]
- Parida, A.K.; Das, A.B. Salt tolerance and salinity effects on plants: A review. Ecotoxicol. Environ. Saf. 2005, 60, 324–349. [Google Scholar] [CrossRef] [PubMed]
- Galvan-Ampudia, C.S.; Testerink, C. Salt stress signals shape the plant root. Curr. Opin. Plant Biol. 2011, 14, 296–302. [Google Scholar] [CrossRef]
- Shannon, M.C. Whole-plant response to salinity. In Plant Environment Interactions; Wilkinson, R.E., Ed.; Marcel Dekker: New York, NY, USA, 1994; pp. 199–244. [Google Scholar]
- Liang, W.; Ma, X.; Wan, P.; Liu, L. Plant salt-tolerance mechanism: A review. Biochem. Biophys. Res. Commun. 2018, 495, 286–291. [Google Scholar] [CrossRef]
- Acosta-Motos, J.R.; Ortuño, M.F.; Bernal-Vicente, A.; Diaz-Vivancos, P.; Sanchez-Blanco, M.J.; Hernandez, J.A. Plant responses to salt stress: Adaptive mechanisms. Agronomy 2017, 7, 18. [Google Scholar] [CrossRef]
- Paganová, V.; Jureková, Z.; Lichtnerová, H. The nature and way of root adaptation of juvenile woody plants Sorbus and Pyrus to drought. Environ. Monit. Assess. 2019, 191, 714. [Google Scholar] [CrossRef]
- Chaves, M.M.; Osorio, J.; Pereira, J.S. Water use efficiency and photosynthesis. In Water Use Efficiency in Plant Biology; Bacon, M., Ed.; Blackwell Publishing: Oxford, UK, 2004; pp. 42–74. [Google Scholar]
- Soltabayeva, A.; Ongaltay, A.; Omondi, J.O.; Srivastava, S. Morphological, Physiological and Molecular Markers for Salt-Stressed Plants. Plants 2021, 10, 243. [Google Scholar] [CrossRef]
- Sun, Y.H.; Shi, R.; Zhang, X.H.; Chiang, V.L.; Sederoff, R.R. MicroRNAs in trees. Plant. Mol. Biol. 2012, 80, 37–53. [Google Scholar] [CrossRef]
- Barvkar, V.T.; Pardeshi, V.C.; Kale, S.M.; Qiu, S.; Rollins, M.; Datla, R.; Gupta, V.S.; Kadoo, N.Y. Genome-wide identification and characterization of microRNA genes and their targets in flax (Linum usitatissimum): Characterization of flax miRNA genes. Planta 2013, 237, 1149–1161. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R. MicroRNAs with macro-effects on plant stress responses. Semin. Cell Dev. Biol. 2010, 21, 805–811. [Google Scholar] [CrossRef]
- Gao, Z.; Ma, C.; Zheng, C.; Yao, Y.; Du, Y. Advances in the regulation of plant salt-stress tolerance by, miRNA. Mol. Biol. Rep. 2022, 49, 5041–5055. [Google Scholar] [CrossRef]
- Kumar, V.; Khare, T.; Shriram, V.; Wani, S.H. Plant small RNAs: The essential epigenetic regulators of gene expression for salt-stress responses and tolerance. Plant Cell Rep. 2018, 37, 61–75. [Google Scholar] [CrossRef] [PubMed]
- Bej, S.; Basak, J. MicroRNAs: The Potential Biomarkers in Plant Stress Response. Am. J. Plant Sci. 2014, 5, 748–759. [Google Scholar] [CrossRef]
- Heng, W.; Jia, B.; Huang, H.; Yang, Y.; Wang, Z.; Liu, P.; Ye, Z.; Zhu, L. Identification of russet-associated microRNAs in the exocarp of a Dangshansuli pear mutant (Pyrus bretschneideri Rehd.) by high-throughput sequencing. Tree Genet. Genomes 2016, 12, 107. [Google Scholar] [CrossRef]
- Liebsch, D.; Palatnik, J.F. MicroRNA miR396, GRF transcription factors and GIF co-regulators: A conserved plant growth regulatory module with potential for breeding and biotechnology. Curr. Opin. Plant Biol. 2020, 53, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Tsukaya, H. Regulation of plant growth and development by the Grothw-Regulatin Factor and GRF-Interacting factor duo. J. Exp. Bot. 2015, 66, 6093–6107. [Google Scholar] [CrossRef]
- Rodriguez, R.E.; Ercoli, M.F.; Debernardi, J.M.; Palatnik, J.F. Growth-Regulating Factors, a Transcription Factor Family Regulating More than just Plant Growth; Gonzalez, D.H., Ed.; Plant Transcription Factors; Academic Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Casadevall, R.; Rodriguez, R.E.; Debernardi, J.M.; Palatnik, J.F.; Casati, P. Repression of growth regulating factors by the microRNA396 inhibits cell proliferation by UV-B radiation in Arabidopsis leaves. Plant Cell 2013, 25, 3570–3583. [Google Scholar] [CrossRef]
- Yuan, S.; Zhao, J.; Li, Z.; Hu, Q.; Yuan, N.; Zhou, M.; Xia, X.; Noorai, R.; Saski, C.; Li, S.; et al. MicroRNA396-mediated alteration in plant development and salinity stress response in creeping bentgrass. Hortic. Res. 2019, 6, 48. [Google Scholar] [CrossRef]
- Gao, P.; Bai, X.; Yang, L.; Lv, D.; Li, Y.; Cai, H.; Ji, W.; Guo, D.; Zhu, Y. Over-expression of osa-MIR396c decreases salt and alkali stress tolerance. Planta 2010, 231, 991–1001. [Google Scholar] [CrossRef] [PubMed]
- Hao, K.; Wang, Y.; Zhu, Z.; Wu, Y.; Chen, R.; Zhang, L. miR160: An Indispensable Regulator in Plant. Front. Plant Sci. 2022, 13, 833322. [Google Scholar] [CrossRef]
- Ma, C.H.; Burd, S.; Lers, A. miR408 is involved in abiotic stress responses in Arabidopsis. Plant J. 2015, 84, 169–187. [Google Scholar] [CrossRef]
- Liu, J.N.; Ma, X.; Yan, L.; Liang, Q.; Fang, H.; Wang, C.; Dong, Y.; Chai, Z.; Zhou, R.; Bao, Y.; et al. MicroRNA and Degradome Profiling Uncover Defense Response of Fraxinus velutina Torr. to Salt Stress. Front. Plant Sci. 2022, 13, 847853. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Li, X.; Deng, M.; Zhao, Z.; Yang, L. Comparative Analysis and Identification of miRNAs and Their Target Genes Responsive to Salt Stress in Diploid and Tetraploid Paulownia fortunei Seedlings. PLoS ONE 2016, 11, e0149617. [Google Scholar] [CrossRef]
- Hansen, C.C.; Nelson, D.R.; Møller, B.L.; Werck-Reichhart, D. Plant cytochrome P450 plasticity and evolution. Mol. Plant. 2021, 14, 1244–1265. [Google Scholar] [CrossRef]
- He, Y.; Yan, H.; Hua, W.; Huang, Y.; Wang, Z. Selection and validation of reference genes for quantitative real-time PCR in Gentiana macrophylla. Front Plant Sci. 2016, 7, 945. [Google Scholar] [CrossRef]
- Ferradás, Y.; Rey, L.; Martínez, Ó.; Rey, M.; González, M.V. Identification and validation of reference genes for accurate normalization of real-time quantitative PCR data in kiwifruit. Plant Physiol Bioch. 2016, 102, 27–36. [Google Scholar] [CrossRef]
- Rumlow, A.; Keunen, E.; Klein, J.; Pallmann, P.; Riemenschneider, A.; Cuypers, A.; Papenbrock, J. Quantitative expression analysis in Brassica napus by Northern blot analysis and reverse transcription-quantitative PCR in a complex experimental setting. PLoS ONE 2016, 11, e0163679. [Google Scholar] [CrossRef]
- Zhang, J.; Gai, M.; Xue, B.; Jia, N.; Wang, C.; Wang, J.; Sun, H. The use of miRNAs as reference genes for miRNA expression normalization during Lilium somatic embryogenesis by real-time reverse transcription PCR analysis. Plant Cell Tissue Organ Cult. 2017, 129, 105–118. [Google Scholar] [CrossRef]
- Bharati, R.; Sen, M.K.; Kumar, R.; Gupta, A.; Žiarovská, J.; Fernández-Cusimamani, E.; Leuner, O. Systematic Identification of Suitable Reference Genes for Quantitative Real-Time PCR Analysis in Melissa officinalis L. Plants 2023, 12, 470. [Google Scholar] [CrossRef]
- Shivhare, R.; Lata, C. Seletion of suitable reference genes for assessing gene expression in pearl millet under different abiotic stresses and their combinations. Sci. Rep. 2016, 6, 23036. [Google Scholar] [CrossRef]
- Qi, X.; Wu, J.; Wang, L.; Li, L.; Cao, Y.; Tian, L.; Dong, X.; Zhang, S. Identifying the candidate genes involved in the calyx abscission process of ‘Kuerlexiangli’ (Pyrus sinkiangensis Yu) by digital transcript abundance measurements. BMC Genom. 2013, 14, 727. [Google Scholar] [CrossRef]
- Wang, Y.Z.; Dai, M.S.; Zhang, S.J.; Shi, Z.B. Exploring candidate genes for pericarp russet pigmentation of sand pear (Pyrus pyrifolia) via RNA-seq data in two genotypes contrasting for pericarp color. PLoS ONE 2014, 9, e83675. [Google Scholar] [CrossRef]
- Yang, S.L.; Zhang, X.N.; Lu, G.L.; Wang, C.R.; Wang, R. Regulation of gibberellin on gene expressions related with the lignin biosynthesis in ‘Wangkumbae’ pear (Pyrus pyrifolia Nakai) fruit. Plant Growth Regul. 2015, 76, 127–134. [Google Scholar] [CrossRef]
- Wu, T.; Zhang, R.; Gu, C.; Wu, J.; Wan, H.; Zhang, S.; Zhang, S. Evaluation of candidate reference genes for real time quantitative PCR normalization in pear fruit. Agric. J. Agr. Res. 2012, 7, 3701–3704. [Google Scholar]
- Imai, T.; Ubi, B.E.; Saito, T.; Moriguchi, T. Evaluation of reference genes for accurate normalization of gene expression for real time-quantitative PCR in Pyrus pyrifolia using different tissue samples and seasonal conditions. PLoS ONE 2014, 9, e86492. [Google Scholar] [CrossRef]
- Chen, J.; Li, X.; Wang, D.; Li, L.; Zhou, H.; Liu, Z.; Wu, J.; Wang, P.; Jiang, X.; Fabrice, M.R.; et al. Identification and testing of reference genes for gene expression analysis in pollen of Pyrus bretschneideri. Sci. Hortic. 2015, 190, 43–56. [Google Scholar] [CrossRef]
- Dai, S.; Li, P.; Chen, P.; Li, Q.; Pei, Y.; He, S.; Sun, Y.; Wang, Y.; Kai, W.; Zhao, B.; et al. Transcriptional regulation of genes encoding ABA metabolism enzymes during the fruit development and dehydration stress of pear ‘Gold Nijisseiki’. Plant Physiol. Biochem. 2014, 82, 299–308. [Google Scholar] [CrossRef]
- Yang, X.; Hu, H.; Yu, D.; Sun, Z.; He, X.; Zhang, J.; Chen, Q.; Tian, R.; Fan, J. Candidate resistant genes of sand pear (Pyrus pyrifolia Nakai) to Alternaria alternata revealed by transcriptome sequencing. PLoS ONE 2015, 10, e0135046. [Google Scholar] [CrossRef]
- Cao, Y.; Han, Y.; Li, D.; Lin, Y.; Cai, Y. MYB transcription factors in Chinese pear (Pyrus bretschneideri Rehd.): Genome-wide identification, classification, and expression profiling during fruit development. Front. Plant Sci. 2016, 7, 577. [Google Scholar] [CrossRef]
- Nham, N.T.; Macnish, A.J.; Zakharov, F.; Mitcham, E.J. ‘Bartlett’ pear fruit (Pyrus communis L.) ripening regulation by low temperatures involves genes associated with jasmonic acid, cold response, and transcription factors. Plant Sci. 2017, 260, 8–18. [Google Scholar] [CrossRef]
- Yao, G.; Ming, M.; Allan, A.C.; Gu, C.; Li, L.; Wu, X.; Wang, R.; Chang, Y.; Qi, K.; Zhang, S.; et al. Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J. 2017, 92, 437–451. [Google Scholar] [CrossRef]
- Sun, K.; Fang, H.; Chen, Y.; Zhuang, Z.; Chen, Q.; Shan, T.; Khan, M.K.R.; Zhang, J.; Wang, B. Genome-Wide Analysis of the Cytochrome P450 Gene Family Involved in Salt Tolerance in Gossypium hirsutum. Front. Plant Sci. 2021, 12, 685054. [Google Scholar] [CrossRef]
- Mao, G.; Seebeck, T.; Schrenker, D.; Yu, O. CYP709B3, a cytochrome P450 monooxygenase gene involved in salt tolerance in Arabidopsis thaliana. BMC Plant Biol. 2013, 13, 169. [Google Scholar] [CrossRef]
- Xu, F.; Peng, M.; Luo, Q.; Jiang, M.; Zhang, X.; Zong, X.; Meng, F.; Li, Y. Isolation and detection of transcript-derived fragments (TDFs) in NaCl-stressed black locust (Robinia pseudoacacia L.) using cDNA-AFLP analysis. Acta Physiol. Plant. 2015, 37, 168. [Google Scholar] [CrossRef]









| Primer | Sequences (5′-3′) |
|---|---|
| lus-miR168_F | CAC GCA TCG CTT GGT GCA GGT |
| lus-miR168_R | CCA GTG CAG GGT CCG AGG TA |
| lus-miR408a_F | GGC TGG GAA CAG ACA GAG CAT GGA |
| lus-miR408a_R | GGG AAA AAG GCC AGG GAA GAG G |
| cca-miR156a_F | TGA CAG AAG AGA GTG AGC AC |
| cca-miR156a_R cca-miR396a_F | GTG CTC ACT CTC TTC TGT CA TTC CAC AGC TTT CTT GAA CTT |
| cca-miR396a_R gb-miR482_F | GTT CAA GAA AGC TGT GGG AAA TGG GTT GTA GTC TTC AGG AGT GGG |
| gb-miR482_R | GAA GGC AAT AGG AAT GGG AGG ATC |
| mdo-miR398_F | TGT GTT CTC AGG TCA GGG GTT |
| mdo-miR398_R | AAC CCC TGA CCT GAG AAC ACA |
| mdo-miR160a_F | TGC CTG GCT CCC TGT ATG CCA |
| mdo_miR160a_R | TGG CAT ACA GGG AGC CAG GCA |
| mdo-miR172a_F | AGA ATC TTG ATG ATG CTG CAT |
| mdo-miR172a_R | ATG CAG CAT CAT CAA GAT TCT |
| Primer Name | Primer Sequence |
|---|---|
| PC Act F1/R1 | F:CTCCCAGGGCTGTGTTTCCTA |
| R:CTCCATGTCATCCCAGTTGCT | |
| PC P450 F1/R1 | F:GAACTCTTGAGGCACCCGAA |
| R:AATGGGGCAACTGGGTGTAG |
| Parameter | p-Value | Control | 100 mM |
|---|---|---|---|
| Stem length (mm) | 0.0024 | 232.75 (±58.67) a | 160.33 (±43.83) b |
| Stem increment (mm) | 0.0023 | 139.25 (±46.04) a | 85.83 (±27.75) b |
| LA (mm2) | 0.0245 | 11,876.10 (±4301.85) a | 8216.16 (±3011.44) b |
| SLA (mm2·mg−1) | 0.7016 | 16.57 (±1.61) a | 16.80 (±1.40) a |
| DW (mg) | 0.0037 | 1426.75 (±483.07) a | 893.50 (±302.38) b |
| DWL (mg) | 0.0114 | 717.92 (±241.29) a | 484.50 (±166.32) b |
| DWS (mg) | 0.0023 | 708.83 (±258.55) a | 409.00 (±154.36) b |
| LWC (%) | 0.1228 | 60.47 (±1.92) a | 61.59 (±1.33) a |
| RL (mm) | 0.1374 | 7312.79 (±1999.09) a | 6321.61 (±752.54) a |
| SRL (mm·mg−1) | 0.1705 | 9.54 (±2.10) a | 8.37 (±1.94) a |
| RSA (mm2) | 0.1707 | 7459.93 (±1191.91) a | 6711.47 (±1213.18) a |
| RV (mm3) | 0.1686 | 2424.01 (±726.21) a | 2054.15 (±480.23) a |
| ARD (mm) | 0.8199 | 0.42 (±0.05) a | 0.41 (±0.04) a |
| NORT | 0.7285 | 1793.00 (±290.89) a | 1852.83 (±490.04) a |
| DWR (mg) | 0.5940 | 740.27 (±169.35) a | 705.46 (±129.51) a |
| R:S | 0.0057 | 0.59 (±0.20) a | 0.90 (±0.28) b |
| Fine root volume (mm3) | 0.1044 | 853.83 (±168.90) a | 720.92 (±203.09) a |
| Volume of coarse roots (mm3) | 0.4846 | 1394.99 (±347.51) a | 1272.78 (±436.85) a |
| Volume of very fine roots 0–1 (mm3) | 0.0350 | 575.95 (±152.73) a | 460.03 (±92.82) b |
| Fine-to-coarse root ratio | 0.4532 | 0.62 (±0.11) a | 0.56 (±0.25) a |
| CSI (mL.Plant−1) | Parameter | p-Value | gs (mmol H2O m−2 s−1) | p-Value | An (μmol CO2 m−2 s−1) | p-Value | E (mmol H2O m−2 s−1) | p-Value | WUE (mmol CO2 mol−1 H2O) |
|---|---|---|---|---|---|---|---|---|---|
| 0 | Control | 0.9736 | 63.00 (±37.36) a | 0.7003 | 5.00 (±2.24) a | 0.5976 | 0.63 (±0.31) a | 0.1277 | 9.36 (±3.91) a |
| 100 mM | 62.50 (±20.91) a | 5.34 (±1.42) a | 0.69 (±0.19) a | 7.34 (±1.17) a | |||||
| 100 | Control | 0.1389 | 57.43 (±31.93) a | 0.2885 | 5.55 (±1.54) a | 0.3598 | 0.57 (±0.30) a | 0.8752 | 10.77 (±5.26) a |
| 100 mM | 35.29 (±18.62) a | 4.69 (±1.50) a | 0.42 (±0.24) a | 11.16 (±4.27) a | |||||
| 175 | Control | 0.0662 | 110.57 (±29.90) a | 0.1467 | 7.01 (±1.37) a | 0.0712 | 1.11 (±0.24) a | 0.9962 | 6.57 (±2.06) a |
| 100 mM | 78.71 (±29.07) a | 5.60 (±1.99) a | 0.85 (±0.26) a | 6.57 (±1.06) a | |||||
| 250 | Control | 0.0758 | 105.14 (±32.92) a | 0.2742 | 5.79 (±1.71) a | 0.0676 | 1.21 (±0.30) a | 0.6642 | 4.90 (±1.37) a |
| 100 mM | 75.38 (±26.86) a | 4.76 (±1.75) a | 0.92 (±0.27) a | 5.17 (±1.01) a | |||||
| 300 | Control | 0.0143 | 44.5 (±12.82) a | 0.0166 | 4.94 (±1.26) a | 0.0178 | 0.56 (±0.23) a | 0.2482 | 9.66 (±3.10) a |
| 100 mM | 24.13 (±13.43) b | 3.02 (±1.31) b | 0.28 (±0.16) b | 11.95 (±3.91) a |
| Stress/Primer Combination | CYPA1F+R * | CYP2BF+R | CYP2CF+R | ||
|---|---|---|---|---|---|
| Length range of amplified fragments | leaves | control before | 30–900 bp | 40–1000 bp | 80–1000 bp |
| 20/100 mL | |||||
| 35/175 mL | |||||
| 60/300 mL | |||||
| control after | |||||
| roots | control before | 180–700 bp | 90–1000 bp | 50–1200 bp | |
| 20/100 mL | 170–1000 bp | ||||
| 35/175 mL | 210–1000 bp | ||||
| 60/300 mL | 170–1000 bp | ||||
| control after | 90–1000 bp | ||||
| Type of obtained profile | leaves | control before | all monomophic | ||
| 20/100 mL | |||||
| 35/175 mL | |||||
| 60/300 mL | |||||
| control after | |||||
| roots | control before | monomorphic | |||
| 20/100 mL | monomorphic | polymorphic | monomorphic | ||
| 35/175 mL | monomorphic | polymorphic | monomorphic | ||
| 60/300 mL | monomorphic | polymorphic | monomorphic | ||
| control after | monomorphic | ||||
| Unique fragments | leaves | control before | no | ||
| 20/100 mL | |||||
| 35/175 mL | |||||
| 60/300 mL | |||||
| control after | |||||
| roots | control before | no | no | no | |
| 20/100 mL | no | yes/220, 790 bp insertion | no | ||
| 35/175 mL | no | yes/220 bp insertion | no | ||
| 60/300 mL | no | yes/670 bp insertion | no | ||
| control after | no | no | no | ||
| Difference in PBA profile of leaves/roots | - | - | no | yes | no |
| Difference in PBA profile of control/stress variants | - | - | no | yes | no |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paganová, V.; Hus, M.; Lichtnerová, H.; Žiarovská, J.; Moravčíková, D.; Kučka, M.; Ražná, K.; Abbas, A. Physiological and Molecular Responses of Pyrus pyraster Seedlings to Salt Treatment Analyzed by miRNA and Cytochrome P450 Gene-Based Markers. Plants 2024, 13, 261. https://doi.org/10.3390/plants13020261
Paganová V, Hus M, Lichtnerová H, Žiarovská J, Moravčíková D, Kučka M, Ražná K, Abbas A. Physiological and Molecular Responses of Pyrus pyraster Seedlings to Salt Treatment Analyzed by miRNA and Cytochrome P450 Gene-Based Markers. Plants. 2024; 13(2):261. https://doi.org/10.3390/plants13020261
Chicago/Turabian StylePaganová, Viera, Marek Hus, Helena Lichtnerová, Jana Žiarovská, Dagmar Moravčíková, Matúš Kučka, Katarína Ražná, and Aqsa Abbas. 2024. "Physiological and Molecular Responses of Pyrus pyraster Seedlings to Salt Treatment Analyzed by miRNA and Cytochrome P450 Gene-Based Markers" Plants 13, no. 2: 261. https://doi.org/10.3390/plants13020261
APA StylePaganová, V., Hus, M., Lichtnerová, H., Žiarovská, J., Moravčíková, D., Kučka, M., Ražná, K., & Abbas, A. (2024). Physiological and Molecular Responses of Pyrus pyraster Seedlings to Salt Treatment Analyzed by miRNA and Cytochrome P450 Gene-Based Markers. Plants, 13(2), 261. https://doi.org/10.3390/plants13020261