Multiple Mobile mRNA Signals Regulate Tuber Development in Potato
Abstract
:1. Introduction
1.1. Phloem-Mobile mRNAs in Plants
1.2. Signals for Potato Tuberization
1.3. The Tuberization Clade of the StBEL Family
2. Mobile RNAs of Potato
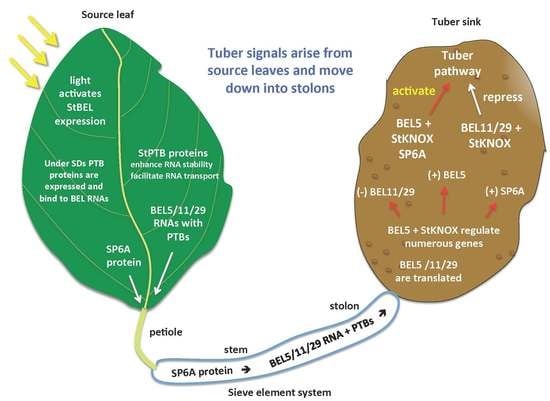
2.1. StBEL5 Functions as a Mobile RNA Signal
2.2. Mobility and Function of StBEL11 and StBEL29
3. Mechanism for StBEL RNA Movement: The Role of the PTB Proteins
4. Final Perspectives
Acknowledgments
Conflicts of Interest
References
- Lucas, W.J.; Groover, A.; Lichtenberger, R.; Furuta, K.; Yadav, S.R.; Helariutta, Y.; He, X.Q.; Fukuda, H.; Kang, J.; Brady, S.M.; et al. The plant vascular system: Evolution, development and functions. J. Integr. Plant Biol. 2013, 55, 294–388. [Google Scholar] [CrossRef] [PubMed]
- Asano, T.; Masumura, T.; Kusano, H.; Kikuchi, S.; Kurita, A.; Shimada, H.; Kadowaki, K. Construction of a specialized cDNA library from plant cells isolated by laser capture microdissection: Toward comprehensive analysis of the genes expressed in the rice phloem. Plant J. 2002, 32, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Vilaine, F.; Palauqui, J.C.; Amselem, J.; Kusiak, C.; Lemoine, R.; Dinant, S. Towards deciphering phloem: A transcriptome analysis of the phloem of Apium graveolens. Plant J. 2003, 36, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Omid, A.; Keilin, T.; Glass, A.; Leshkowitz, D.; Wolf, S. Characterization of phloem-sap transcription profile in melon plants. J. Exp. Bot. 2007, 58, 3645–3656. [Google Scholar] [CrossRef] [PubMed]
- Deeken, R.; Ache, P.; Kajahn, I.; Klinkenberg, J.; Bringmann, G.; Hedrich, R. Identification of Arabidopsis thaliana phloem RNAs provides a search criterion for phloem-based transcripts hidden in complex datasets of microarray experiments. Plant J. 2008, 55, 746–759. [Google Scholar] [CrossRef] [PubMed]
- Gaupels, F.; Buhtz, A.; Knauer, T.; Deshmukh, S.; Waller, F.; van Bel, A.J.E.; Kogel, K.H.; Kehr, J. Adaptation of aphid stylectomy for analyses of proteins and mRNAs in barley phloem sap. J. Exp. Bot. 2008, 59, 3297–3306. [Google Scholar] [CrossRef] [PubMed]
- Notaguchi, M.; Higashiyama, T.; Suzuki, T. Identification of mRNAs that move over long distances using an RNA-Seq analysis of Arabidopsis/Nicotiana benthamiana heterografts. Plant Cell Physiol. 2015, 56, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Thieme, C.J.; Rojas-Triana, M.; Stecyk, E.; Schudoma, C.; Zhang, W.; Yang, L.; Miñambres, M.; Walther, D.; Schulze, W.X.; Paz-Ares, J.; et al. Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat. Plants 2015, 1, 15025. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.K.; Chatterjee, M.; Yu, Y.; Suh, S.G.; Miller, W.A.; Hannapel, D.J. Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. Plant Cell 2006, 18, 3443–3457. [Google Scholar] [CrossRef] [PubMed]
- Ghate, T.H.; Sharma, P.; Khondare, K.R.; Hannapel, D.J.; Banerjee, A.K. The mobile RNAs, StBEL11 and StBEL29, suppress growth of tubers in potato. Plant Mol. Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, A.; Bhogle, S.; Kang, I.H.; Hannapel, D.J.; Banerjee, A.K. The mRNA of a Knotted1-like transcription factor of potato is phloem mobile. Plant Mol. Biol. 2012, 79, 595–608. [Google Scholar] [CrossRef] [PubMed]
- Haywood, V.; Yu, T.S.; Huang, N.C.; Lucas, W.J. Phloem long-distance trafficking of GIBBERELLIC ACID-INSENSITIVE RNA regulates leaf development. Plant J. 2005, 42, 49–68. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Canio, W.; Kessler, S.; Sinha, N. Developmental changes due to long distance movement of a homeobox fusion transcript in tomato. Science 2001, 293, 287–289. [Google Scholar] [CrossRef] [PubMed]
- Notaguchi, M.; Wolf, S.; Lucas, W.J. Phloem-mobile Aux/IAA transcripts target to the root tip and modify root architecture. J. Integr. Plant Biol. 2012, 54, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Gu, M.; Shi, N.; Zhang, H.; Yang, X.; Osman, T.; Liu, Y.; Wang, H.; Vatish, M.; Jackson, S.; et al. Mobile FT mRNA contributes to the systemic florigen signalling in floral induction. Sci. Rep. 2011, 1, 73. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.C.; Jane, W.N.; Chen, J.; Yu, T.S. Arabidopsis CENTRORADIALIS homologue acts systemically to inhibit floral initiation in Arabidopsis. Plant J. 2012, 72, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.J.; Huang, N.C.; Liu, Y.S.; Lu, C.A.; Yu, T.S. Long-distance movement of Arabidopsis FLOWERING LOCUS T RNA participates in systemic floral regulation. RNA Biol. 2012, 9, 653–662. [Google Scholar] [CrossRef] [PubMed]
- Molnar, A.; Melnyk, C.W.; Bassett, A.; Hardcastle, T.J.; Dunn, R.; Baulcombe, D.C. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 2010, 328, 872–875. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Marshall, D.; Bryan, G.J.; Hornyik, C. Identification and characterization of miRNA transcriptome in potato by high-throughput sequencing. PLoS ONE 2013, 8, e57233. [Google Scholar] [CrossRef] [PubMed]
- Lakhotia, N.; Joshi, G.; Bhardwaj, A.R.; Katiyar-Agarwal, S.; Agarwal, M.; Jagannath, A.; Goel, S.; Kumar, A. Identification and characterization of miRNAome in root, stem, leaf and tuber developmental stages of potato (Solanum tuberosum L.) by high-throughput sequencing. BMC Plant Biol. 2014, 14, 6. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.; Adam, H.; Díaz-Mendoza, M.; Zurczak, M.; González-Schain, N.D.; Suárez-López, P. Graft-transmissible induction of potato tuberization by the microRNA miR172. Development 2009, 136, 2873–2881. [Google Scholar] [CrossRef] [PubMed]
- Bhogale, S.; Mahajan, A.S.; Natarajan, B.; Rajabhoj, M.; Thulasiram, H.V.; Banerjee, A.K. MicroRNA156: A potential graft-transmissible microRNA that modulates plant architecture and tuberization in Solanum tuberosum ssp. andigena. Plant Physiol. 2014, 164, 1011–1027. [Google Scholar] [CrossRef] [PubMed]
- Kanehira, A.; Yamada, K.; Iwaya, T.; Tsuwamoto, R.; Kasai, A.; Nakazono, M.; Harada, T. Apple phloem cells contain some mRNAs transported over long distances. Tree Genet. Genomes 2010, 5, 635–642. [Google Scholar] [CrossRef]
- Ham, B.K.; Brandom, J.L.; Xoconostle-Cazares, B.; Ringgold, V.; Lough, T.L.; Lucas, W.J. A polypyrimidine tract binding protein, pumpkin RBP50, forms the basis of a phloem-mobile ribonucleoprotein complex. Plant Cell 2009, 21, 197–215. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Medrano, R.; Xoconostle-Cazares, B.; Lucas, W.J. Phloem long-distance transport of CmNACP mRNA: Implications for supracellular regulation in plants. Development 1999, 126, 4405–4419. [Google Scholar] [PubMed]
- Yang, H.W.; Yu, T.S. Arabidopsis floral regulators FVE and AGL24 are phloem-mobile RNAs. Bot. Stud. 2010, 51, 17–26. [Google Scholar]
- Navarro, C.; Abelenda, J.A.; Cruz-Oró, E.; Cuéllar, C.A.; Tamaki, S.; Silva, J.; Shimamoto, K.; Prat, S. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 2011, 478, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Kloosterman, B.; Abelenda, J.A.; del Mar Gomez, M.; Oortwijn, M.; de Boer, J.M.; Kowitwanich, K.; Horvath, B.M.; van Eck, H.J.; Smaczniak, C.; Prat, S.; et al. Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature 2013, 495, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Lin, T.; Hannapel, D.J. Targets of the StBEL5 transcription factor include the FT ortholog StSP6A. Plant Physiol. 2016, 170, 310–324. [Google Scholar] [CrossRef] [PubMed]
- Bürglin, T.R. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucl. Acids Res. 1997, 25, 4173–4180. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Sharma, P.; Gonzalez, D.H.; Viola, I.L.; Hannapel, D.J. The impact of the long-distance transport of a BEL1-like mRNA on development. Plant Physiol. 2013, 161, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Rutjens, B.; Bao, D.; van Eck-Stouten, E.; Brand, M.; Smeekens, S.; Proveniers, M. Shoot apical meristem function in Arabidopsis requires the combined activities of three BEL1-like homeodomain proteins. Plant J. 2009, 58, 641–654. [Google Scholar] [CrossRef] [PubMed]
- Ung, N.; Lal, S.; Smith, H.M. The role of PENNYWISE and POUND-FOOLISH in the maintenance of the shoot apical meristem in Arabidopsis. Plant Physiol. 2011, 156, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.M.S.; Hake, S. The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell 2003, 15, 1717–1727. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Tabb, P.; Hepworth, S.R. BLADE-ON-PETIOLE1 and 2 regulate Arabidopsis inflorescence architecture in conjunction with homeobox genes KNAT6 and ATH1. Plant Signal Behav. 2012, 7, 788–792. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.; Wang, Y.; Franzen, R.; Santi, L.; Salamini, F.; Rohde, W. In vitro interactions between barley TALE proteins suggest a role for protein-protein associations in the regulation of Knox gene function. Plant J. 2001, 27, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Banerjee, A.K.; Hannapel, D.J. The tandem complex of BEL and KNOX partners is required for transcriptional repression of ga20ox1. Plant J. 2004, 38, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Pagnussat, G.C.; Yu, H.J.; Sundaresan, V. Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1. Plant Cell 2007, 19, 3578–3592. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Lin, T.; Grandellis, C.; Yu, M.; Hannapel, D.J. The BEL1-like family of transcription factors in potato. J. Exp. Bot. 2014, 65, 709–723. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Rosin, F.M.; Hannapel, D.J. Interacting transcription factors from the three amino acid loop extension superclass regulate tuber formation. Plant Physiol. 2003, 132, 1391–1404. [Google Scholar] [CrossRef] [PubMed]
- Abelenda, J.A.; Navarro, C.; Prat, S. Flowering and tuberization: A tale of two nightshades. Trends Plant Sci. 2014, 19, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Suárez-López, P. A critical appraisal of phloem-mobile signals involved in tuber induction. Front. Plant Sci. 2013, 4, 253. [Google Scholar] [CrossRef] [PubMed]
- Hannapel, D.J. A perspective on photoperiodic phloem-mobile signals that control development. Front. Plant Sci. 2013, 4, 295. [Google Scholar] [CrossRef] [PubMed]
- Bellaoui, M.; Pidkowich, M.S.; Samach, A.; Kushalappa, K.; Kohalmi, S.E.; Modrusan, Z.; Crosby, W.L.; Haughn, G.W. The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell 2001, 13, 2455–2470. [Google Scholar] [CrossRef] [PubMed]
- Brambilla, V.; Battaglia, R.; Colombo, M.; Masiero, S.; Bencivenga, S.; Kater, M.M.; Colombo, L. Genetic and molecular interactions between BELL1 and MADS box factors support ovule development in Arabidopsis. Plant Cell 2007, 19, 2544–2556. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.K.; Lin, T.; Hannapel, D.J. Untranslated regions of a mobile transcript mediate RNA metabolism. Plant Physiol. 2009, 151, 1831–1843. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.K.; Sharma, P.; Butler, N.M.; Kang, I.H.; Shah, S.; Rao, A.G.; Hannapel, D.J. Polypyrimidine tract-binding proteins of potato mediate tuberization through an interaction with StBEL5 RNA. J. Exp. Bot. 2015, 66, 6835–6847. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, M.; Banerjee, A.K.; Hannapel, D.J. A BELL1-like gene of potato is light activated and wound inducible. Plant Physiol. 2007, 145, 1435–1443. [Google Scholar] [CrossRef] [PubMed]
- Ayre, B.G.; Blair, J.E.; Turgeon, R. Functional and phylogenetic analyses of a conserved regulatory program in the phloem of minor veins. Plant Physiol. 2003, 133, 1229–1239. [Google Scholar] [CrossRef] [PubMed]
- Kloosterman, B.; Navarro, C.; Bijsterbosch, G.; Lange, T.; Prat, S.; Visser, R.G.; Bachem, C.W. StGA2ox1 is induced prior to stolon swelling and controls GA levels during potato tuber development. Plant J. 2007, 52, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Carrera, E.; Bou, J.; García-Martínez, J.L.; Prat, S. Changes in GA 20-oxidase gene expression strongly affect stem length, tuber induction and tuber yield of potato plants. Plant J. 2000, 22, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Eviatar-Ribak, T.; Shalit-Kaneh, A.; Chappell-Maor, L.; Amsellem, Z.; Eshed, Y.; Lifschitz, E. A cytokinin-activating enzyme promotes tuber formation in tomato. Curr. Biol. 2013, 23, 1057–1064. [Google Scholar] [CrossRef] [PubMed]
- Rosin, F.M.; Hart, J.K.; van Onckelen, H.; Hannapel, D.J. Suppression of a vegetative MADS box gene of potato activates axillary meristem development. Plant Physiol. 2003, 131, 1613–1622. [Google Scholar] [CrossRef] [PubMed]
- Roumeliotis, E.; Kloosterman, B.; Oortwijn, M.; Visser, R.G.; Bachem, C.W. The PIN family of proteins in potato and their putative role in tuberization. Front. Plant Sci. 2013, 14, 524. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Cao, X.; Shi, S.; Ma, Y.; Wang, K.; Liu, S.; Chen, D.; Chen, Q.; Ma, H. Genome-wide survey of Aux/IAA gene family members in potato (Solanum tuberosum): Identification, expression analysis, and evaluation of their roles in tuber development. Biochem. Biophys. Res. Commun. 2016. [Google Scholar] [CrossRef] [PubMed]
- Kolomiets, M.V.; Hannapel, D.J.; Chen, H.; Tymeson, M.; Gladon, R.J. Lipoxygenase is involved in the control of potato tuber development. Plant Cell 2001, 13, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.C.; Yu, T.S. The sequences of Arabidopsis GA-INSENSITIVE RNA constitute the motifs that are necessary and sufficient for RNA long-distance trafficking. Plant J. 2009, 59, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Corral-Debrinski, M.; Blugeon, C.; Sawicka, J.C. In yeast, the 3′untranslated region or the presequence of ATM1 is required for the exclusive localization of its mRNA to the vicinity of mitochondria. Mol. Cell. Biol. 2000, 20, 7881–7892. [Google Scholar] [CrossRef] [PubMed]
- Thio, G.L.; Ray, R.P.; Barcelo, G.; Schüpbach, T. Localization of gurken RNA in Drosophila oogenesis requires elements in the 5′ and 3′ regions of the transcript. Dev. Biol. 2000, 221, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.P. mRNA localization: Message on the move. Nat. Rev. Mol. Cell. Biol. 2001, 2, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Sawicka, K.; Bushell, M.; Spriggs, K.A.; Willis, A.E. Polypyrimidine tract-binding protein: A multifunctional RNA-binding protein. Biochem. Soc. Trans. 2008, 36, 641–647. [Google Scholar] [CrossRef] [PubMed]
- King, M.L.; Messitt, T.J.; Mowry, K.L. Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol. Cell. 2005, 97, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Eliscovich, C.; Buxbaum, A.R.; Katz, Z.B.; Singer, R.H. mRNA on the move: The road to its biological destiny. J. Biol. Chem. 2013, 288, 20361–20368. [Google Scholar] [CrossRef] [PubMed]
- Buxbaum, A.R.; Haimovich, G.; Singer, R.H. In the right place at the right time: Visualizing and understanding mRNA localization. Nat. Rev. Mol. Cell. Biol. 2015, 16, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Auweter, S.D.; Allain, F.H.T. Structure–function relationships of the polypyrimidine tract binding protein. Cell. Mol. Life Sci. 2008, 65, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Vreugdenhil, D.; van Lammeren, A.A.M. Cell division and cell enlargement during potato tuber formation. J. Exp. Bot. 1998, 49, 573–582. [Google Scholar] [CrossRef]
- Xu, X.; van Lammeren, A.A.M.; Vermeer, E.; Vreugdenhil, D. The role of gibberellin, abscisic acid, and sucrose in the regulation of potato tuber formation in vitro. Plant Physiol. 1998, 117, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Sanz, M.J.; Mingo-Castel, A.; van Lammeren, A.A.M.; Vreugdenhil, D. Changes in the microtubular cytoskeleton precede in vitro tuber formation in potato. Protoplasma 1996, 191, 46–54. [Google Scholar] [CrossRef]
- Shibaoka, H. Regulation by gibberellins of the orientation of cortical microtubules in plant cells. Aust. J. Plant Physiol. 1993, 20, 461–470. [Google Scholar] [CrossRef]
- Roumeliotis, E.; Kloosterman, B.; Oortwijn, M.; Kohlen, W.; Bouwmeester, H.J.; Visser, R.G.; Bachem, C.W. The effects of auxin and strigolactones on tuber initiation and stolon architecture in potato. J. Exp. Bot. 2012, 63, 4539–4547. [Google Scholar] [CrossRef] [PubMed]
- Kolachevskaya, O.O.; Alekseeva, V.V.; Sergeeva, L.I.; Rukavtsova, E.B.; Getman, I.A.; Vreugdenhil, D.; Buryanov, Y.I.; Romanov, G.A. Expression of auxin synthesis gene tms1 under control of a tuber-specific promoter enhances potato tuberization in vitro. J. Integr. Plant Biol. 2015, 57, 734–744. [Google Scholar] [CrossRef] [PubMed]
- Bolduc, N.; Yilmaz, A.; Mejia-Guerra, M.K.; Morohashi, K.; O’Connor, D.; Grotewold, E.; Hake, S. Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Develop. 2012, 26, 1685–1690. [Google Scholar] [CrossRef] [PubMed]
- Lifschitz, E.; Ayre, B.G.; Eshed, Y. Florigen and anti-florigen—A systemic mechanism for coordinating growth and termination in flowering plants. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Hanzawa, Y.; Money, T.; Bradley, D. A single amino acid converts a repressor to an activator of flowering. Proc. Natl. Acad. Sci. USA 2005, 102, 7748–7753. [Google Scholar] [CrossRef] [PubMed]








| RNA | Annotation | Putative Function | Reference |
|---|---|---|---|
| MpSLR/IAA14 | Auxin response factor | Transcriptional repressor | [23] |
| CmSCL14P | Scarecrow-like | Transcription factor | [24] |
| CmSTM | Shoot meristemless | Meristem regulator | [24] |
| CmERF | Ethylene response factor | Ethylene signaling | [24] |
| CmNAC | NAM, ATAF1/2 and CUC2 | Meristem development | [25] |
| CmMyb | Myb-like transcription factor | Transcriptional activator | [24] |
| BoFVE | Mammalian retinoblastoma-associated protein | Floral regulator | [26] |
| BoAGL24 | Agamous-like | Floral regulator | [26] |
| AtAux/IAA18 and -28 * | Auxin response factor | Auxin signaling ⇩ | [14] |
| CmGAI * | GA Insensitive | Leaf morphology ⇧ | [12] |
| StBEL5 * | Potato BEL1-like family | Tuber growth ⇩ | [9] |
| StBEL11/29 * | Potato BEL1-like family | Tuber growth ⇩ | [10] |
| POTH1 * | Potato KNOTTED1-type | Vegetative growth ⇩ | [11] |
| PFP-LeT6 * | Tomato Knotted1-type fusion | Leaf morphology ⇧ | [13] |
| FT * | Arabidopsis Flowering locus T | Activates flowering ⇧ | [15] |
| ATC * | Arabidopsis CENTRORADIALIS | Represses flowering ⇧ | [16] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hannapel, D.J.; Banerjee, A.K. Multiple Mobile mRNA Signals Regulate Tuber Development in Potato. Plants 2017, 6, 8. https://doi.org/10.3390/plants6010008
Hannapel DJ, Banerjee AK. Multiple Mobile mRNA Signals Regulate Tuber Development in Potato. Plants. 2017; 6(1):8. https://doi.org/10.3390/plants6010008
Chicago/Turabian StyleHannapel, David J., and Anjan K. Banerjee. 2017. "Multiple Mobile mRNA Signals Regulate Tuber Development in Potato" Plants 6, no. 1: 8. https://doi.org/10.3390/plants6010008
APA StyleHannapel, D. J., & Banerjee, A. K. (2017). Multiple Mobile mRNA Signals Regulate Tuber Development in Potato. Plants, 6(1), 8. https://doi.org/10.3390/plants6010008