1. Introduction
The spread of invasive alien species (IAS) has received a great deal of attention, given the adverse ecological and economic effects that they can impose on invaded ecosystems [
1,
2]. Why invasive species expand their range faster in non-native regions, but not in their native range, and how these species become dominant components of non-native habitats while being minor constituents of their native ecosystems [
3] are still open questions. Alien plants are those plants that have been introduced to an area from their native range, either accidentally or intentionally, whereas invasive plants are those plants that are non-native to an ecosystem and which may cause a threat to native biodiversity and ecosystem integrity in many ecologically sensitive parts of the world. Very recently, the International Union for Conservation of Nature has come up with a standard Environmental Impact Classification of Alien Taxa (EICAT), based on the magnitude of the detrimental environmental impacts. Earlier studies have indicated that invasive species often possess specific demographic or physiological traits, such as faster growth rates, greater productivity, high fecundity, greater tolerance to environmental extreme events, or very effective dispersal compared to native and non-invasive introduced species [
4,
5,
6,
7]. These traits enable introduced species to spread very extensively and achieve high abundance. The origin of such variations in traits that influence success in competition between invasive and native species under natural conditions may include geographic isolation, genetic drift, divergent selection, phenotypic plasticity [
8], and rapid adaptive evolution [
9,
10]. Whether phenotypic divergence among and between populations of alien species is due to one or more of these factors is a debate that remains to be resolved.
The phenotypic differences within and among populations of invasive species in their native and non-native ranges are widely speculated [
11,
12]. The colonization of new areas by invasive species would be associated theoretically with founder diversity [
13], phenotypic plasticity, and environmental conditions at the points of origin [
14], various ecological and evolutionary processes associated with geographical zone [
15], and population bottlenecks that reduce within-population genetic diversity and increase genetic differentiation among populations [
16]. These population genetic differentiations have important effects on species invasiveness, which is apparently linked to increased genetic variation and evolutionary potential [
17]. Moreover, population genetic analyses can increase our understanding of the origin and expansion of invasive species and invasive populations [
18,
19], or haplotypes (i.e., individuals with a specific group of alleles that are inherited within a species), and the same could have useful management implications as well. Previous research has typically focused on detecting how invasive haplotypes have invaded and replaced native haplotypes [
20,
21]. This replacement is most often due to better performance of invasive haplotypes compared to native haplotypes with respect to morphological traits, such as plant height and size, and to reproductive success. However, little attention has been paid to understanding the variation patterns of such invasive haplotypes in their native regions. Further, the causes of morphological differences between invasive and natural populations of IAS are not well understood. Thus, investigating phenotypic and genotypic differentiation between invasive and native haplotypes in wild populations should contribute to the understanding of mechanisms behind the invasion of alien species.
Among-population phenotypic differentiation in the wild is effectively estimated using
PST (i.e., the phenotypic analogue of
QST); [
22,
23,
24]. Lande (1992) [
25] stated that genetic variance in phenotypic traits between populations is expected to equal that of variation in neutral molecular loci under the influence of the forces that drive evolution. Indeed, the level of differentiation of quantitative traits should be similar to the level of differentiation at neutral loci, as divergent selection in quantitative traits induces more morphological differences than would be expected for neutral molecular markers [
24]. For this reason, a valuable method for investigating morphological differences between invasive and natural populations of IAS is to compare divergence in allele frequencies to divergence in quantitative traits. Whether selection or genetic drift is responsible for phenotypic divergence is a complex task and requires the initial step of comparing genetic differentiation with phenotypic differentiation. Phenotypic differences between populations are often assumed to be adaptive, and epigenetics are reported to play key role on adaptation of invasive species [
26]. Both molecular and quantitative variations can be influenced by non-adaptive processes that involve random genetic drift and gene flow [
27,
28]. Under these conditions, it would be especially challenging and interesting to determine whether phenotypic differences between populations represent the outcomes of adaptive or non-adaptive processes.
Common reed (
Phragmites australis [Cav.] Trin. ex Steud; Poaceae) is a polyploid perennial grass species with clonal architecture [
29,
30,
31]. The species is distributed worldwide and is highly invasive in Kashmir Himalayan aquatic habitats [
32]. In Kashmir, India, this species was earlier reported as a South American native [
33] and European native [
32], but substantial molecular evidence for the nativity of this species is lacking. Saltonstall (2002) [
29] sampled individuals of
P. australis worldwide and recovered 27 haplotypes of this species on the basis of the sequences of two chloroplast DNA (hereafter, cpDNA) fragments. In North America, both native and exotic haplotypes of the species are found. Introduction of an exotic genotype of
P. australis (Haplotype M) and its rapid spread, aided by anthropogenic disturbance, has contributed significantly to its invasion and range expansion in North America [
29]. Recently, Guo et al. [
34] showed that evolutionary mechanisms act differently in the native and introduced ranges of
P. australis and that invasive populations of European origin have evolved to adapt to a different climate and to human-made habitats in North America. In Quebec, Canada, the native haplotype is characterized by a red stem and ligule base (i.e., the junction of the leaf blade with the leaf sheath) with fewer hairs, compared to the green stem of invasive haplotypes, which have more hairs on the ligule. Moreover, the inflorescence of the native haplotype is less dense relative to that of the invasive haplotype, containing fewer seeds [
35,
36]. The invasive haplotype in Quebec tends to grow preferentially along roads and in other human-made habitats, while the native haplotype prefers undisturbed, pristine habitats [
37,
38,
39,
40]. In India, the invasive
P. australis haplotype is found to occur widely in various aquatic habitats, especially in Kashmir Himalayan wetlands.
The objectives of this study were: (i) to assess and compare the phenotypic and genetic diversity of invasive populations of P. australis in Kashmir and Quebec, together with native haplotypes of this species in Quebec, and (ii) to assess evolutionary similarities between them.
2. Results
Restriction fragment length polymorphism (hereafter, RFLP) analysis confirmed that there was only one native population (QN1) in our sampled populations, instead of five as thought based on morphological variations. In utilizing the cpDNA PCR-RFLP analyses, we found that the Quebec population QN1 shared the same restriction pattern as native populations in USA [
41]. Further, we found that ten populations (QE1 to QE12) exhibited the same restriction sites as haplotype M [
42]. Results for the sequence
trnT-
trnL in the native Quebec QN1 population was 99% similar to haplotype E2 (NCBI accession no. AY016325; [
42]). Sequence
trnT-
trnL in the invasive Quebec QE1 population and sequence
trnT-
trnL in the Kashmir KE1 population were 99% similar to haplotype 4 (NCBI accession no. AY016327; [
42]). BLAST analysis was done in GenBank NCBI database for the
rbcL-
psaI intergenic spacer, and for native Quebec QN1, the sequence was identical to haplotype E2 (100% identity with NCBI accession no. JQ409547; Freeland and Vachon Unpublished data). Sequence
rbcL-
psaI in the invasive Quebec QE1 population was 99% similar to haplotype 4, while Kashmir KE1 population was identical (100% similar) to the same sequence of haplotype 4 (NCBI accession no. AY016335, [
42]). All nine populations from Kashmir displayed the same restriction pattern as haplotype M. Based on this information, we considered the putative native Quebec populations of Lac St-Louis, Parc Louis-Racine, Sablière Colette, and La Pocatière as invasive
P. australis populations, referring to them instead as QE9, QE10, QE11, and QE12.
2.1. Phenotypic Diversity
The invasive
P. australis plants of the Ange-Gardien population, Quebec (QE4) were the tallest (Mean ± SE: 230.74 ± 55.54 cm height), followed by the invasive haplotypes of Bernière, Quebec (QE8: 226.72 ± 34.63 cm height), and the invasive haplotypes of Eastman, Quebec (QE6: 212.44 ± 68.96 cm height;
Table 1). The thickest plant stems were recorded for invasive
P. australis plants from Kuhunus Wullar populations (KE5: 7.83 ± 1.362 cm), Ashaibagh (KE2: 7.78 ± 1.332 cm) and Rangharstop (KE4: 7.74 ± 1.264 cm; Table 5). The heaviest flowers in terms of fresh and dry mass were also recorded in Ashaibagh, Ange-Gardien, and Bernière populations (
Table 1). The highest intermodal length was recorded in plants from Bernière (QE8: 18.00 ± 3.749 cm), La Pocatière (QE1: 16.16 ± 2.407 cm), and Princeville (QE7: 16.77 ± 2.266 cm;
Table 1). The longest leaves were recorded in Ashaibagh, Rangharstop, Ange-Gardien, and Eastmann
P. australis populations, whereas the widest leaves were recorded in invasive
P. australis plants from Kashmir (
Table 1).
Highly significant variation was observed among plants, whereas significant to highly significant variation was found among populations for the measured characters, except for leaf width (
Table 2). The variation between individual plants and between populations most often contributed to the bulk of total variation in the measured characters, except for leaf width (
Table 2). Additionally, among-populations variation was higher than between-haplotypes variation for all the characters that were measured (
Table 2). While
P. australis plants from Kashmir had more highly significant (
p < 0.001) heavier flower dry mass (3.59 ± 0.59 g) than did Quebec plants (2.55 ± 0.116 g), invasive
P. australis plants had significantly wider leaves (2.04 ± 0.036 cm) than did native plants (1.65 ± 0.05 cm). Additionally, except for flower and dry seeds mass, no variation was found among regions for all measured characters (
Table 2).
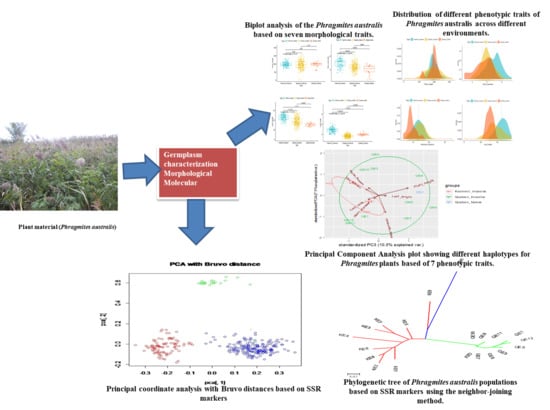
2.2. Characterization of Phenotypes
To investigate the effects of morphological traits in distinguishing the haplotypes that were investigated, we analyzed the distribution of each trait among haplotypes. Though stem diameter, plant height, leaf length, and width distinguished the haplotypes that were studied, the differences were not significant (
Figure 1).
To explore descriptors, we measured seven phenotypes (plant height, stem diameter, fresh flowers, dry flowers, internode length, and leaf length and width) in
P. australis. A bimodal distribution was observed for stem diameter, internode length, leaf length, and width (
Figure 2). Normal distributions without any significant skewness were observed for the rest of traits.
Principal component analysis (PCA) indicated two principal components with eigen-values greater than 1. The PCA grouped the seven
P. australis descriptors into various components, with the first two explaining 79.33% of the variation, while the first principal axis (PC1) alone explained 61.84% of the variation (
Figure 3). The PC1 had high loading for leaf width. The second axis (PC2), explaining about 17.49% of the total variation, was correlated with plant height, stem diameter, fresh flowers and seeds, dry flowers, and seeds, internode length, and leaf length, indicating the usefulness of descriptors in
P. australis (
Table 3).
2.3. Phenotypic Differences and Heritability
Moderate
PST estimates were observed among populations, ranging from 0.12 for flower and seed fresh mass to 0.41 for leaf length (
Table 2). The measured traits were weakly (leaf width) to moderately (internode length, leaf length, and flower masses) heritable (
Table 2).
2.4. Genetic Diversity
The polymorphism information content (PIC) varied from 0.4 (PaGT11) to 0.9, with an average of 0.8 (
Table 4). Based on the number of alleles, it was found that
P. australis in Kashmir shows hexaploidy as the number of alleles from each population ranged from 1–6; for Quebec populations, it shows tetraploidy as the number of alleles ranged from 1–4. However, Quebec native population plants seem to be diploid, having only 1–2 alleles (
Table 4). All loci that were surveyed were variable in each population. Within-population genetic diversity indices are summarized in
Table 5. The mean number of alleles per locus ranged from 1.89 to 4.56, and the overall mean number of alleles per locus amounted to 8.56. The mean number of alleles per polymorphic locus ranged from 1.61 to 3.37, whereas the overall mean number of alleles per polymorphic locus amounted to 4.16 (
Table 5). For
P. australis populations in Kashmir,
A ranged from 2.00 to 4.56 and
Ap ranged from 2.04 to 3.37. The range of values for
A and
Ap was not as wide in the Quebec populations, and respectively, 1.89 to 3.56, and 1.61 to 2.32. Mean percentages of polymorphic loci ranged from 66.67% (KE9, Ganderbal, Kashmir) to 100% (KE1, KE4, KE5, KE6, KE7, and KE8) for Kashmir populations. For the Quebec populations,
P ranged from 88.89% (QE1, QE3, QE6, QE8, QE9, QE12, and QN1) to 100% (QE4, QE7, QE10, and QE11). The overall percentage of polymorphic loci was 92.78% for all study populations. Allelic richness for Kashmiri and Quebec populations ranged from 1.81 to 2.54, and 1.54 to 2.18, respectively. Observed and expected heterozygosities for Kashmir populations were 0.70 and 0.54, respectively; for Quebec populations,
HO was 0.38 and
HE was 0.39. Overall mean observed and expected estimates of heterozygosity were 0.52 and 0.46, respectively, while
HO and
HE per population ranged from 0.23 (QN1; Lac St François, Quebec) to 0.78 (KE6; Saderkote wullar, Kashmir) and from 0.20 (QN) to 0.66 (KE6), respectively (
Table 5).
2.5. Population Genetic Structure
High genetic differentiation was observed among populations (overall
RST = 0.353), ranging from 0.133 between invasive populations from Kashmir and Quebec, to 0.418 between Quebec native and invasive populations. These differences were best illustrated by the results of principal coordinate analysis (
Figure 4). The
P. australis plants separated into three groups: Kashmir invasive, Quebec natives, and Quebec invasive, which each formed one group (
Figure 4).
The phylogenetic tree (
Figure 5) was congruent with principal coordinate analysis results (
Figure 4), as the populations of native and non-native
P. australis that were studied were found to be very structured. Clearly, ten populations belonging to Quebec invasive (QE1, QE2, QE4, QE6, QE7, QE8, QE9, QE10, QE11, and QE12) were grouped together, completely distant from the population of Quebec native (QN1;
Figure 5). Kashmir invasive populations were clustered in the same group, whereas KE9 were close to QN1 population following by a sub-group containing height populations (KE1, KE2, KE3, KE4, KE5, KE6, KE7, and KE8;
Figure 5).
2.6. RST- PST Comparisons
Among-population phenotypic differentiation estimates for plant height (0.23), plant stem diameter (0.25), and fresh and dry mass of flowers (0.12 and 0.14, respectively) were less than overall RST (0.35), whereas PST estimate for leaf width (0.39) and leaf length (0.41) exceeded the genetic differentiation at nine cpDNA microsatellite loci among the P. australis populations that were surveyed in the present study. PST estimate for internode length (0.33) was comparable with among-population RST estimate (0.35).
2.7. Relationships between Genetic and Geographic Distances
All the populations from both regions revealed a weak positive relationship between genetic distances and environmental variables (r2 = 0.389, p < 0.001). When Mantel test was calculated within each region it again showed a weak but significant correlation between the two variables in Quebec (r2 = 0.158, p ≤ 0.05) and non-significant correlation in Kashmir (r2 = 8.06 × 10−3, p = 0.5570).
3. Discussion
The behavior of
P. australis in various lakes, wetlands, and other water bodies of Kashmir is that of a typical invader in terms of forming the monotypic stands, attaining great abundance, and outcompeting native species [
43,
44]. Previous studies that were carried out in 2018 [
45] based on field observations reported this species at the stage V (widespread and dominant) on the stage-based hierarchical model (CM model) propounded by Colautti and MacIsaac [
46]. Similarly, earlier studies conducted in 2007 [
33] reported this species as a South American native and Shah et al. [
32] in 2014 reported it as a European native based on published, web-based sources as well as phytogeographical distribution of this species, but substantial molecular evidence for the nativity of this species is lacking. Based on the above evidence, many authors declared that
P. australis was introduced in India, especially in Kashmir Himalayan region [
44,
45]. There is hardly any study conducted in Kashmir, India, or elsewhere, that suggests native range of
P. australis in this part (Kashmir Himalaya) of Asia. The present study aimed at increasing our understanding of the phenotypic and genetic diversity of invasive populations of
P. australis in Kashmir and Quebec. Only one native
P. australis population in Quebec was identified during field surveys on reported natural distribution of the species across Quebec province and all the populations from Kashmir turned out to be invasive as revealed by cpDNA PCR-RFLP analyses. Such findings indicate that invasive haplotypes are becoming more dominant in the natural distribution range of the species in the sites of study. This is the first study in India, especially in Kashmir Himalaya, that provides molecular evidence regarding the invasive status of
P. australis in this region. Such successful invasion of alien haplotypes in their non-native range is due to the fact that invaders usually perform better than natives in terms of life-history traits such as mature plant size and fecundity, thereby reducing the size of, or even replacing, native populations [
4,
7]. Further studies in this direction need to be carried out by taking samples from a larger global range, including native European populations of
P. australis, in order to provide greater understanding of the origin and phylogeography of this species.
3.1. Phenotypic Variation
Given the premise that the invasive populations perform better than native ones in terms of various growth-related traits, we found the highest values of seven growth-related traits of
P. australis within invasive populations. These results are congruent with that of other studies [
47,
48,
49], although DeVrie et al. [
50] indicated that morphological characters cannot discriminate reliably between invasive and native
P. australis individuals in North America. Further, local environment highly affects phenotypic traits in common reed [
51], explaining the difficulty in differentiating invasive and native individuals from these traits. However, plasticity in common reed is trait-specific [
51,
52]. Similarly, Pysek et al. [
53] compared the functional traits of 89 populations in a common garden experiment belonging to distinct clones of different origin: North America, Europe, Australia (two groups including native and invasive populations introduced from Europe), South Africa, and Far East. In general, both native and invasive populations with origins in Europe showed highest performance-related traits than other groups. North American native populations performed more poorly than both European-related groups and were similar in this respect to Australian and South African populations [
53]. Invasive European populations in North America do suppress the native North American populations wherever growing in direct competition [
54]. The weaker performance of Australian and South African populations in the Northern Hemisphere is due to weak genetic differences or environmental similarities [
53].
Plant height plays an important role in invasiveness, as Crawley et al. [
55] and Canavan et al. [
56] found that tall-statured grasses harbor a useful functional trait for invasion success. In their review, Mozdzer et al. [
57] clearly indicated that introduced and native
P. australis lineages differ both physiologically and morphologically. Introduced plants are generally taller and occur in greater densities, which results in greater productivity in the introduced lineage in nearly every study.
In the present study, plant stem diameter appeared to be linked to invasiveness of
P. australis, given that the highest mean values were recorded for invasive populations. Yet Muth and Pigliucci [
58], in comparing six growth-related traits, found very little variation among introduced invasive and introduced non-invasive plant species within two genera of Asteraceae (
Crepis and
Centaurea), with respect to stem diameter. As our study included one native population, we suggest that any assumptions need to be confirmed with fine-scale studies over a broader distribution range of
P. australis, taking into account an even larger number of populations.
Abundant flowering may also play an important role in reproductive success, which obviously influences propagule pressure and invasiveness. Our results that invasive plants in both regions significantly produce more flowers and seeds than native plants are quite striking. These traits can be taken as the indicator/s of plant invasiveness. In a survey of 46 native and 45 introduced populations of
Solidago gigantea, Jakobs et al. [
59] found that total plant biomass was larger for invasive than native plants. Some genetic studies have shown that abundant seed production and seed-based recruitment are more common in
P. australis compared to dispersal by stolons and rhizomes [
60,
61,
62]. Albert et al. [
63], however, attributed the success of this invader in North America to a reproduction strategy combining both the advantages of generative and vegetative propagations.
Comparing the leaf characteristics of invasive and native populations showed that the former exhibit higher values for leaf area than the latter. Bimodal distribution observed for traits related to stem diameter, internode distance, leaf length, and width suggest that a major gene controls those four important characters related to leaf and stem sizes in
P. australis. Although there was not significant among-population variation in leaf width, results from the present study revealed maximum leaf length and leaf width for invasive populations, while the lowest values were encountered in native population. Furthermore, variation in leaf width was very highly significant between haplotypes in the present study, with invaders appearing to perform better with respect to this quantitative trait as well. Successful invaders are characterized by a wider range of leaf areas and by greater phenotypic plasticity [
64]. Several studies have shown that invasive species have higher resource capture-related traits and possess more efficient photosynthetic machinery compared to native or non-invasive species [
11,
65,
66,
67]. Invasiveness is strongly related to leaf traits that are associated with rapid C capture and net CO
2 assimilation [
68,
69] and relative growth rate [
70].
3.2. Characterization of Phenotypes
The characterization of among-haplotype variation could provide clues on co-selection of traits in case hitchhiking occurs [
71]. Indeed, a group of characters may distinguish closely related taxa, which are geographically distant, because of genetic drift or hitchhiking. Similar effects are also observed when a major gene controls the characters of interest. Results of the present study revealed that at least stem diameter, leaf length, and width may undergo co-selection or the traits are under the control of the major gene. Genome-wide association studies in the species may provide more insights on the issue.
3.3. Genotypic Studies
After obtaining useful insights into phenotypic trait-based variations in native and invasive populations’ vis-à-vis invasiveness in
P. australis, our objective was to confirm such variations at molecular level. Results from the present study indicate that in general, highest levels of genetic diversity are found in invasive populations, while the lowest levels of diversity are recorded in native population, consistent with [
72]. The highest levels of genetic diversity in invasive populations may explain adaptability of invaders in non-native habitats [
9,
10]. The low levels of genetic diversity in the only one native population that was identified in the present study are congruent with the results of [
73], which observed lower levels of genetic diversity in native
P. australis populations than in invaders at many of cpDNA loci. This is contrary to what was observed by Zhao et al. [
74], who reported highest genetic diversity among native Mexican populations of
Solanum rostratum Dunal and significantly reduced diversity in invasive populations within the USA and China, using ten microsatellite markers. Since cpDNA PCR-RFLP haplotype identification analyses in the present study revealed only one native population, instead of five as thought on the basis of morphological variations, and all the other populations are exotic, it seems that invasion has either replaced the native populations or intraspecific hybridization might have occurred between native and introduced lineages of a species that has increased invasiveness and lead to the decline of native lineages [
54]. We suggest that a broad-scale study including more continents than what has been done in our study, after identifying more native populations, be carried out to confirm or refute our findings.
Being a cryptic invader,
P. australis has emerged as an important model system for understanding mechanisms of invasion. There have been few population-level studies to directly evaluate differences in genetic diversity, reproductive mode, and dispersal between native and non-native
P. australis, particularly in western North America [
62]. In view of the lack of any population genetics studies on
P. australis in India, we have attempted here to assess its genetic diversity through allelic and population variation of microsatellite loci. It is noteworthy that the mean number of alleles per locus in the present study (
A = 8.56) was comparable to the findings of Saltonstall [
73] on European
P. australis populations (
A = 8.0), although it was greater than the average (
A = 7.6) as reported by Paul et al. [
75] on
P. australis in the United Kingdom. In the present study, we most frequently came across tetraploidy and hexaploidy among regional groupings of invasive populations of
P. australis that were collected across Quebec and Kashmir, although the native population in Quebec seems to be diploid. Hence, it appears that polyploidy might have played some role in invasiveness of this species. Clevering and Lissner [
76] found that tetraploidy was the most common ploidy of
P. australis in Europe and North America, while octaploids (8x) were predominated in Asia. Such results suggest that invasive plants are more likely to be polyploids, while native plants are more likely to be diploids as was reported in some other studies [
77,
78]. Pandit et al. [
79] showed that invasive plant species are generally found to have high chromosome counts and to be polyploids, in contrast to rare species that are diploids and have lower ploidy ratios. In a related study, it was shown that the highly invasive plant species in Singapore were all polyploids [
80], thereby indicating that polyploidy contributes to invasion success. Interestingly, invasive Kashmir and invasive Quebec populations of
P. australis were more similar to one another, both in terms of the number of alleles and fragment size, than to the native Quebec population that showed unique numbers of alleles and fragment sizes. With an increased native population size, more differences might be seen. However, Saltonstall (2003a) [
30] reported the same allelic variation among native
P. australis, while highest allelic diversity was observed among introduced
P. australis that were collected in North America.
Our results indicate that
P. australis populations that were studied are structured, which is in agreement with Guo et al. [
34]. While the latter study revealed the presence of substructures to delineate the population structure in common reed, Guo et al. [
81] postulated that the climatic niche had shifted between the native and introduced populations of the invasive European lineage. The authors reported strong effects of geography and environment on the genetic structure of populations in the native range, but these effects were counteracted by human-related factors manifested through colonization of anthropogenic habitats in the introduced range. Another possible explanation of the substructures observed in our study may be borrowed from Albert et al. [
63], who while investigating the reproduction of
P. australis in eastern North America proposed that long-distance seed dispersal is imperative for common reed along roadsides and in marshes; whereas, plant fragments as well as seeds were shown to be important for dispersal to short distances along roads at least in the areas of its abundance. Further, robust reproductive strategy, both through sexual and asexual modes, was shown to mainly contribute to the invasive success of this species in North America. Our phylogenetic analysis easily identified three mains groups, with each group showing clear specificity in terms of the investigated traits.
3.4. Phenotypic vs. Genotypic Differentiation
Our results suggest that environmental conditions in Quebec and Kashmir are not as much contrasting to induce divergent selection on quantitative traits through strong local population pressures [
82]. This is because there was no major variation in genetic structure between Quebec and Kashmir
P. australis populations, and no significant phenotypic variation was recorded between Quebec and Kashmir populations, except for flower and seed dry mass. Such non-spatial geographic differences in phenotypic traits are not common and may be caused by probable multiple introductions [
17,
83]. However, difference in flower and seed dry mass between the regions indicates that there might be differential effect of selection on quantitative characters.
The among-population phenotypic divergence that was observed in this study regardless of region is not surprising and may be explained by several factors, including plasticity, genetic drift, and divergent selection. As
RST differed to
PST for six out of seven studied characters, divergent selection appears as the most probable cause of among-populations difference in quantitative traits [
84]. No apparent physical barrier to gene flow existed between sampled populations within the region, suggesting that genetic barriers such as different ploidy levels or the high rate of local lateral vegetative propagation of the species may explain the reduced gene flow among populations. Results from the present study are in contrast with that of [
30], which investigated patterns of differentiation in nine microsatellite loci in
P. australis. Whether the observed phenotypic divergence in
P. australis populations is heritable is not known. Investigating fine-scale local spatial invasions (i.e., within geographic regions) would certainly shed more light on the possible contributions of plasticity to patterns of among-population phenotypic variation in
P. australis.
The
RST-
PST contrast is useful for evaluating the relative contributions of non-adaptive and adaptive processes in shaping quantitative traits. Local adaptation and evolutionary potential have been advocated as greatly influencing the outcomes of invasion by alien species [
10,
17]. Heritability estimates of the characters that were studied may provide clues on local adaptation. Results of the present study indicate local adaptation of invasive haplotypes for internode length, leaf length, and flower masses, indicating that these traits may influence invasion success in the species. On the other hand, isolation by distance or divergent selection are the main hypotheses that explain phenotypic divergence in nature within species [
8]. As we observed a contrast between
RST and
PST estimates for six out of the seven traits that were investigated, we tested the correlation between genetic distances and geographical distances. Our results showed no or weak relationships between genetic and geographic distances in the regions that were surveyed. Thus, the hypothesis that could explain the phenotypic divergence observed between populations in traits that were investigated in our study is divergent selection for plant height, plant stem diameter, fresh and dry flower mass, leaf length, and leaf width coupled with local adaptation in internode length, leaf length, and flower masses. We also postulate that internode length variation between populations are explained by genetic drift. However, a large fine-scale study would confirm these findings.