Carbohydrate Accumulation and Differential Transcript Expression in Winter Wheat Lines with Different Levels of Snow Mold and Freezing Tolerance after Cold Treatment
Abstract
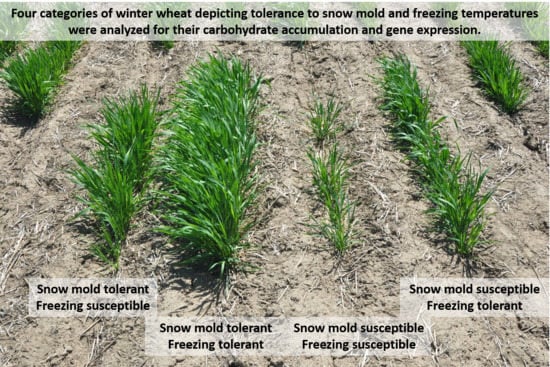
:1. Introduction
2. Results
2.1. Carbohydrate Accumulation
2.2. Transcript Expression of Genes of Interest
2.3. Modular Gene Enrichment
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growing Conditions
4.2. Carbohydrate Extraction and Quantification
4.3. Statistical Analysis
4.4. RNA Extraction, RNAseq, and Modular Enrichment Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| RIL | recombinant inbred line |
| GO | gene ontology |
| BLUPs | best linear unbiased predictions |
| FrT | freezing tolerant |
| FrS | freezing susceptible |
| SMT | snow mold tolerant |
| SMS | snow mold susceptible |
References
- Stelmakh, A.F. Growth habit in common wheat (Triticum aestivum L. EM. Thell.). Euphytica 1987, 36, 513–519. [Google Scholar] [CrossRef]
- Sherman, J.D.; Talbert, L.E. Vernalization-induced changes of the DNA methylation pattern in winter wheat. Genome 2002, 45, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Loukoianov, A.; Blechl, A.; Tranquilli, G.; Ramakrishna, W.; SanMiguel, P.; Bennetzen, J.L.; Echenique, V.; Dubcovsky, J. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 2004, 303, 1640–1644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ergon, Å.; Klemsdal, S.S.; Tronsmo, A.M. Interactions between cold hardening and Microdochium nivale infection on expression of pathogenesis-related genes in winter wheat. Physiol. Mol. Plant Pathol. 1998, 53, 301–310. [Google Scholar] [CrossRef]
- Trischuk, R.G.; Schilling, B.S.; Low, N.H.; Gray, G.R.; Gusta, L.V. Cold acclimation, de-acclimation and re-acclimation of spring canola, winter canola and winter wheat: The role of carbohydrates, cold-induced stress proteins and vernalization. Environ. Exp. Bot. 2014, 106, 156–163. [Google Scholar] [CrossRef]
- Gaudet, D.A.; Laroche, A.; Yoshida, M. Low temperature-wheat-fungal interactions: A carbohydrate connection. Physiol. Plant. 1999, 106, 437–444. [Google Scholar] [CrossRef]
- McBeath, J.H. Snow mold-plant-antagonist interactions: Survival of the fittest under the snow. Plant Health Instr. 2002, 1–4. [Google Scholar] [CrossRef]
- Yoshida, M.; Abe, J.; Moriyama, M.; Kuwabara, T. Carbohydrate levels among winter wheat cultivars varying in freezing tolerance and snow mold resistance during autumn and winter. Physiol. Plant. 1998, 103, 8–16. [Google Scholar] [CrossRef]
- Suzuki, M.; Nass, H.G. Fructan in winter wheat, triticale, and fall rye cultivars of varying cold hardiness. Can. J. Bot. 1988, 66, 1723–1728. [Google Scholar] [CrossRef]
- Tamura, K.I.; Sanada, Y.; Tase, K.; Yoshida, M. Fructan metabolism and expression of genes coding fructan metabolic enzymes during cold acclimation and overwintering in timothy (Phleum pratense). J. Plant Physiol. 2014, 171, 951–958. [Google Scholar] [CrossRef]
- Yukawa, T.; Kobayashi, M.; Watanabe, Y.; Yamamoto, S. Studies on Fructan Accumulation in Wheat (Triticum aestivum L.): IV. Fructan accumulation under cold treatments and its varietal difference in relation to the activities of sucrose-sucrose fructosyl transferase and fructan exohydrolase. Jpn. J. Crop Sci. 1995, 64, 801–806. [Google Scholar] [CrossRef] [Green Version]
- Snider, C.S.; Hsiang, T.; Zhao, G.; Griffith, M. Role of ice nucleation and antifreeze activities in pathogenesis and growth of snow molds. Phytopathology 2000, 90, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Bruehl, G.W.; Cunfer, B. Physiologic and environmental factors that affect the severity of snow mold of wheat. Phytopathology 1971, 61, 792–799. [Google Scholar] [CrossRef]
- Mohammad, F.; Windes, J.M.; Souza, E. Total non-structural carbohydrates in winter wheat populations segregating for snow mold tolerance. Crop Sci. 1997, 37, 108–112. [Google Scholar] [CrossRef]
- Pollock, C.J.; Jones, T. Seasonal patterns of fructan metabolism in forage grasses. New Phytol. 1979, 83, 9–15. [Google Scholar] [CrossRef]
- Kiyomoto, R.K.; Bruehl, G.W. Carbohydrate accumulation and depletion by winter cereals differing in resistance to Typhula idahoensis. Phytopathology 1977, 67, 206–211. [Google Scholar] [CrossRef] [Green Version]
- Kawakami, A.; Yoshida, M. Graminan Breakdown by Fructan Exohydrolase Induced in Winter Wheat Inoculated with Snow Mold. J. Plant Physiol. 2012, 169, 294–302. [Google Scholar] [CrossRef]
- Hisano, H.; Kanazawa, A.; Kawakami, A.; Yoshida, M.; Shimamoto, Y.; Yamada, T. Transgenic Perennial Ryegrass Plants Expressing Wheat Fructosyltransferase Genes Accumulate Increased Amounts of Fructan and Acquire Increased Tolerance on a Cellular Level to Freezing. Plant Sci. 2004, 167, 861–868. [Google Scholar] [CrossRef] [Green Version]
- Ritsema, T.; Smeekens, S. Fructans: Beneficial for Plants and Humans. Curr. Opin. Plant Biol. 2003, 6, 223–230. [Google Scholar] [CrossRef]
- Van den Ende, W.; De Coninck, B.; Van Laere, A. Plant Fructan Exohydrolases: A Role in Signaling and Defense? Trends Plant Sci. 2004, 9, 523–528. [Google Scholar] [CrossRef]
- Zhu, J.; Pearce, S.; Burke, A.; Robert, D.; Daniel, S.; Dubcovsky, J.; Garland, K. Copy number and haplotype variation at the VRN—A1 and central FR—A2 loci are associated with frost tolerance in hexaploid wheat. Theor. Appl. Genet. 2014, 127, 1183–1197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kruse, E.B.; Carle, S.W.; Wen, N.; Skinner, D.Z.; Murray, T.D.; Garland-Campbell, K.A.; Carter, A.H. Genomic regions associated with tolerance to freezing stress and snow mold in winter wheat. G3-Genes Genom. Genet. 2017, 7, 775–780. [Google Scholar] [CrossRef] [Green Version]
- Galiba, G.; Vágújfalvi, A.; Li, C.; Soltész, A.; Dubcovsky, J. Regulatory genes involved in the determination of frost tolerance in temperate cereals. Plant Sci. 2009, 176, 12–19. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, K.; Christov, N.K.; Tsuda, S.; Imai, R. Identification of a novel LEA protein involved in freezing tolerance in wheat. Plant Cell Physiol. 2014, 55, 136–147. [Google Scholar] [CrossRef] [Green Version]
- Holková, L.; Prášil, I.T.; Bradáčová, M.; Vítámvás, P.; Chloupek, O. Screening for frost tolerance in wheat using the expression of dehydrine genes Wcs120 and Wdhn13 at 17 °C. Plant Breed. 2009, 128, 420–422. [Google Scholar] [CrossRef]
- Baek, K.H.; Skinner, D.Z. Alteration of antioxidant enzyme gene expression during cold acclimation of near-isogenic wheat lines. Plant Sci. 2003, 165, 1221–1227. [Google Scholar] [CrossRef]
- Janda, T.; Szalai, G.; Leskó, K.; Yordanova, R.; Apostol, S.; Popova, L.P. Factors contributing to enhanced freezing tolerance in wheat during frost hardening in the light. Phytochemistry 2007, 68, 1674–1682. [Google Scholar] [CrossRef]
- Hiilovaara-Teijo, M.; Palva, E.T. Molecular responses in cold-adapted plants. In Cold-Adapted Organisms; Springer: Berlin/Heidelberg, Germany, 1999; pp. 349–384. [Google Scholar]
- Kosová, K.; Vítámvás, P.; Prášil, I.T. Wheat and barley dehydrins under cold, drought, and salinity–what can LEA-II proteins tell us about plant stress response. Front. Plant Sci. 2014, 5, 343. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhang, H.; Wang, L.; Qian, H.; Qi, X. Extraction of oat (Avena sativa L.) antifreeze proteins and evaluation of their effects on frozen dough and steamed bread. Food Bioprocess Technol. 2015, 8, 2066–2075. [Google Scholar] [CrossRef]
- Koike, M.; Okamoto, T.; Tsuda, S.; Imai, R. A novel plant defensin-like gene of winter wheat is specifically induced during cold acclimation. Biochem. Biophys. Res. Commun. 2002, 298, 46–53. [Google Scholar] [CrossRef]
- Gaudet, D.A.; Laroche, A.; Frick, M.; Huel, R.; Puchalski, B. Cold induced expression of plant defensin and lipid transfer protein transcripts in winter wheat. Physiol. Plant 2003, 117, 195–205. [Google Scholar] [CrossRef]
- Shimada, S.; Ogawa, T.; Kitagawa, S.; Suzuki, T.; Ikari, C.; Shitsukawa, N.; Abe, T.; Kawahigashi, H.; Kikuchi, R.; Handa, H.; et al. A genetic network of flowering-time genes in wheat leaves, in which an APETALA1/FRUITFULL-like gene, VRN1, is upstream of FLOWERING LOCUS. Plant J. 2009, 58, 668–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juhász, Z.; Boldizsár, Á.; Nagy, T.; Kocsy, G.; Marincs, F.; Galiba, G.; Bánfalvi, Z. Pleiotropic effect of chromosome 5A and the mvp mutation on the metabolite profile during cold acclimation and the vegetative/generative transition in wheat. BMC Plant Biol. 2015, 15, 57. [Google Scholar]
- Wang, W.; Wang, X.; Huang, M.; Cai, J.; Zhou, Q.; Dai, T.; Cao, W.; Jiang, D. Hydrogen Peroxide and Abscisic Acid Mediate Salicylic Acid-Induced Freezing Tolerance in Wheat. Front. Plant Sci. 2018, 9, 1137. [Google Scholar] [CrossRef]
- Pour-Benab, S.; Fabriki-Ourang, S.; Mehrabi, A. Expression of dehydrin and antioxidant genes and enzymatic antioxidant defense under drought stress in wild relatives of wheat. Biotechnol. Biotechnol. Equip. 2019, 33, 1063–1073. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, K.; Kuwabara, C.; Umeki, N.; Fujioka, M.; Saburi, W.; Matsui, H.; Abe, F.; Imai, R. The cold-induced defensin TAD1 confers resistance against snow mold and Fusarium head blight in transgenic wheat. J. Biotechnol. 2016, 228, 3–7. [Google Scholar] [CrossRef]
- Hajheidari, M.; Eivazi, A.; Buchanan, B.B.; Wong, J.H.; Majidi, I.; Salekdeh, G.H. Proteomics uncovers a role for redox in drought tolerance in wheat. J. Proteome Res. 2007, 6, 1451–1460. [Google Scholar] [CrossRef]
- Soltész, A.; Tímár, I.; Vashegyi, I.; Tóth, B.; Kellős, T.; Szalai, G.; Vágújfalvi, A.; Kocsy, A.; Galiba, G. Redox changes during cold acclimation affect freezing tolerance but not the vegetative/reproductive transition of the shoot apex in wheat: Redox changes during cold acclimation in wheat. Plant Biol. 2011, 13, 757–766. [Google Scholar] [CrossRef]
- Suzuki, N.; Koussevitzky, S.; Mittler, R.; Miller, G. ROS and redox signalling in the response of plants to abiotic stress: ROS and redox signalling in plants. Plant Cell Environ. 2012, 35, 259–270. [Google Scholar] [CrossRef]
- Kim, J.H.; Chan, K.L.; Mahoney, N.; Campbell, B.C. Antifungal activity of redox-active benzaldehydes that target cellular antioxidation. Ann. Clin. Microbiol. Antimicrob. 2011, 10, 23. [Google Scholar] [CrossRef] [Green Version]
- Carter, C.; Thornburg, R.W. Is the nectar redox cycle a floral defense against microbial attack? Trends Plant Sci. 2004, 9, 320–324. [Google Scholar] [CrossRef] [PubMed]
- Vera-Estrella, R.; Blumwald, E. Plant Defense Response to Fungal Pathogens. Plant Physiol. 1994, 104, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, B.J.; Park, S.; Jameel, M.I.; Kraft, J.C.; Thomashow, M.F.; Schemske, D.W.; Oakley, C.G. Genetic and physiological mechanisms of freezing tolerance in locally adapted populations of a winter annual. Am. J. Bot. 2019, 102, 250–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, K.A.; Allan, R.E.; Anderson, J.; Pritchett, J.A.; Little, L.M.; Morris, C.F.; Line, R.F.; Chen, X.; Walker-Simmons, M.K.; Carter, B.P.; et al. Registration of ‘Finch’ wheat. Crop Sci. 2005, 45, 1656–1657. [Google Scholar] [CrossRef]
- Peterson, C.J.; Allan, R.E.; Rubenthaler, G.L.; Line, R.F. Registration of ‘Eltan’ wheat. Crop Sci. 1991, 31, 1704. [Google Scholar] [CrossRef]
- Balow, K.; Shelton, G.; Burke, A.; Hagemeyer, K.; Klarquist, E.F.; Froese, P.S.; Kruse, E.B.; Carle, S.W.; Roa, A.; Nielson, N.; et al. Registration of the Finch-Eltan winter wheat recombinant inbred line mapping population. J. Plant Reg. 2019, 13, 287–293. [Google Scholar] [CrossRef]
- Gaudet, D.A.; Laroche, A.; Puchalski, B. Seeding date alters carbohydrate accumulation in winter wheat. Crop Sci. 2001, 41, 728–738. [Google Scholar] [CrossRef]
- Ekvall, J.; Stegmark, R.; Nyman, M. Content of low molecular weight carbohydrates in vining peas (Pisum sativum) related to harvest time, size and brine grade. Food Chem. 2006, 94, 513–519. [Google Scholar] [CrossRef]
- nlme: Linear and Nonlinear Mixed Effects Models. Available online: https://cran.r-project.org/web/packages/nlme/nlme.pdf (accessed on 19 October 2020).
- Wu, A.; Neff, N.; Kalisky, T.; Dalerba, P.; Treutlein, B.; Rothenberg, M.; Mburu, F.; Mantalas, G.; Sim, S.; Clarke, S. Quantitative assessment of single-cell RNA-sequencing methods. Nat. Methods 2014, 11, 41–46. [Google Scholar] [CrossRef] [Green Version]
- Griffith, M.; Griffith, O.L.; Mwenifumbo, J.; Goya, R.; Morrissy, A.S.; Morin, R.D.; Corbett, R.; Tang, M.J.; Hou, Y.-C.; Pugh, T.J.; et al. Alternative Expression Analysis by RNA Sequencing. Nat. Methods 2010, 7, 843–847. [Google Scholar] [CrossRef]
- Asmann, Y.W.; Klee, E.W.; Thompson, E.A.; Perez, E.A.; Middha, S.; Oberg, A.L.; Therneau, T.M.; Smith, D.I.; Poland, G.A.; Wieben, E.D.; et al. 3′ Tag Digital Gene Expression Profiling of Human Brain and Universal Reference RNA Using Illumina Genome Analyzer. BMC Genom. 2009, 10, 531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; He, M. Differential Gene Expression Identified by RNA-Seq and QPCR in Two Sizes of Pearl Oyster (Pinctada Fucata). Gene 2014, 538, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [Green Version]
- GO.db: A Set of Annotation Maps Describing the Entire Gene Ontology. Available online: https://bioconductor.org/packages/release/data/annotation/html/GO.db.html (accessed on 19 October 2020).
- SystemsGenetics/FUNC-E v1.0. Zenodo. Available online: https://zenodo.org/record/1246882#.X43pcO17mUk (accessed on 19 October 2020).
- Huang, D.; Sherman, B.T.; Tan, Q.; Collins, J.R.; Alvord, W.G.; Roayaei, J.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. The DAVID gene functional classification tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007, 8, R183. [Google Scholar] [CrossRef] [Green Version]





| Module | SMT/FrT | SMS/FrS | SMT/FrS | SMS/FrT |
|---|---|---|---|---|
| Peroxizome and oxidative stress | 2.2889 | 2.6257 | 6.6852 | |
| Metabolism and redox reactions | 5.6142 | 4.2229 | ||
| Nuclear binding and protein modification | 6.2492 | 4.8104 | ||
| Cell wall carbohydrates and chitin metabolism | 2.6723 | 9.4938 | ||
| Beta-glucan synthesis | 2.0558 |
| Comparisons | SMT/FrT and SMS/FrS | SMT/FrT and SMT/FrS | SMT/FrT and SMS/FrT | SMS/FrS and SMT/FrS | SMS/FrS and SMS/FrT | SMT/FrS and SMS/FrT | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Module Week: | 3 | 11 | 3 | 11 | 3 | 11 | 3 | 11 | 3 | 11 | 3 | 11 |
| Acyl transferase activity | 3.44 | |||||||||||
| Carbohydrate metabolism | 2.62 | |||||||||||
| Cell wall carbohydrates and chitin metabolism | 8.38 | 3.36 | 4.35 | 7.78 | 6.60 | |||||||
| Cellulose synthesis | 5.75 | |||||||||||
| Membrane transport and ion channels | 2.99 | |||||||||||
| Metabolism and redox reactions | 3.42 | 2.69 | 4.35 | 3.09 | 2.99 | 6.81 | ||||||
| mRna modification | 2.69 | |||||||||||
| Nuclear binding and protein modification | 9.16 | 5.87 | 37.67 | 2.88 | ||||||||
| Peroxisome and oxidative stress | 5.21 | 4.57 | ||||||||||
| Category | RIL | SMT Score a | FrT % b |
|---|---|---|---|
| tolerant parent | Eltan | 6.33 | 68 |
| SMT/FrT | FERIL-20 | 7 | 81 |
| FERIL-71 | 6.67 | 85 | |
| SMT/FrS | FERIL-94 | 6 | 15 |
| FERIL-103 | 6 | 8 | |
| SMS/FrT | FERIL-128 | 3.33 | 58 |
| FERIL-132 | 3 | 68 | |
| SMS/FrS | FERIL-74 | 1.67 | 5 |
| FERIL-101 | 2 | 8 | |
| susceptible parent | Finch | 4.33 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kruse, E.B.; Revolinski, S.; Aplin, J.; Skinner, D.Z.; Murray, T.D.; Edwards, C.G.; Carter, A.H. Carbohydrate Accumulation and Differential Transcript Expression in Winter Wheat Lines with Different Levels of Snow Mold and Freezing Tolerance after Cold Treatment. Plants 2020, 9, 1416. https://doi.org/10.3390/plants9111416
Kruse EB, Revolinski S, Aplin J, Skinner DZ, Murray TD, Edwards CG, Carter AH. Carbohydrate Accumulation and Differential Transcript Expression in Winter Wheat Lines with Different Levels of Snow Mold and Freezing Tolerance after Cold Treatment. Plants. 2020; 9(11):1416. https://doi.org/10.3390/plants9111416
Chicago/Turabian StyleKruse, Erika B., Samuel Revolinski, Jesse Aplin, Daniel Z. Skinner, Timothy D. Murray, Charles G. Edwards, and Arron H. Carter. 2020. "Carbohydrate Accumulation and Differential Transcript Expression in Winter Wheat Lines with Different Levels of Snow Mold and Freezing Tolerance after Cold Treatment" Plants 9, no. 11: 1416. https://doi.org/10.3390/plants9111416
APA StyleKruse, E. B., Revolinski, S., Aplin, J., Skinner, D. Z., Murray, T. D., Edwards, C. G., & Carter, A. H. (2020). Carbohydrate Accumulation and Differential Transcript Expression in Winter Wheat Lines with Different Levels of Snow Mold and Freezing Tolerance after Cold Treatment. Plants, 9(11), 1416. https://doi.org/10.3390/plants9111416