Selection of Reference Genes for qRT-PCR Analysis in Medicinal Plant Glycyrrhiza under Abiotic Stresses and Hormonal Treatments
Abstract
:1. Introduction
2. Results
2.1. Expression Profiling of Candidate Reference Genes
2.2. Stability of Candidate Reference Genes
2.3. Optimal Reference Genes Under Different Experimental Conditions
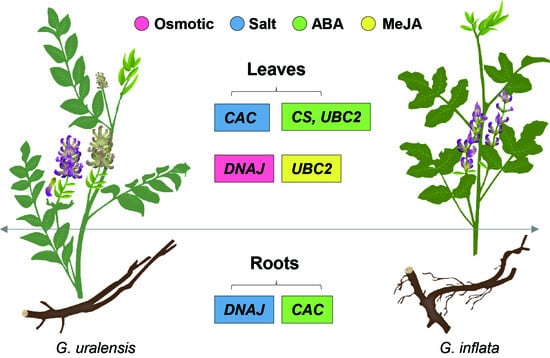
2.4. Comparison of the Suitable Reference Genes under Different Experimental Conditions
2.5. Validation of Recommended Reference Genes
2.6. Summary of Selected Reference Genes within the Leguminosae Plants
3. Discussion
3.1. Stability of Candidate Reference Genes
3.2. Comparison of the Suitable Reference Genes under Different Experimental Conditions
3.3. Suitable Reference Genes for Glycyrrhiza Species under the Osmotic Stress
3.4. Summary of Recommended Reference Genes within the Leguminosae Plants
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Stability Analysis of Candidate Reference Genes
4.3. Comparison of the Suitable Reference Genes under Different Experimental Conditions
4.4. Survey of the Reference Genes Used within the Leguminosae Plant
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Data Availability Statement
References
- Batiha, G.; Alkazmi, L.M.; Wasef, L.G.; Beshbishy, A.M.; Nadwa, E.H.; Rashwan, E.K. Syzygium aromaticum L. (Myrtaceae): Traditional Uses, Bioactive Chemical Constituents, Pharmacological and Toxicological Activities. Biomolecules 2020, 10, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibson, M.R. Glycyrrhiza in old and new perspectives. Lloydia 1978, 41, 348–354. [Google Scholar] [PubMed]
- Fiore, C.; Eisenhut, M.; Krausse, R.; Ragazzi, E.; Pellati, D.; Armanini, D.; Bielenberg, J. Antiviral effects of Glycyrrhiza species. Phytother. Res. PTR 2008, 22, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Cinatl, J.; Morgenstern, B.; Bauer, G.; Chandra, P.; Rabenau, H.; Doerr, H.W. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet 2003, 361, 2045–2046. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Chan, K.H.; Jiang, Y.; Kao, R.Y.; Lu, H.T.; Fan, K.W.; Cheng, V.C.; Tsui, W.H.; Hung, I.F.; Lee, T.S.; et al. In vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2004, 31, 69–75. [Google Scholar] [CrossRef]
- Bailly, C.; Vergoten, G. Glycyrrhizin: An alternative drug for the treatment of COVID-19 infection and the associated respiratory syndrome? Pharmacol. Ther. 2020, 214, 107618. [Google Scholar] [CrossRef]
- Nomura, T.; Fukai, T. Phenolic constituents of licorice (Glycyrrhiza species). Fortschritte der Chemie organischer Naturstoffe = Progress in the chemistry of organic natural products. Prog. Chim. Subst. Org. Nat. 1998, 73, 1–158. [Google Scholar] [CrossRef]
- Mochida, K.; Sakurai, T.; Seki, H.; Yoshida, T.; Takahagi, K.; Sawai, S.; Uchiyama, H.; Muranaka, T.; Saito, K. Draft genome assembly and annotation of Glycyrrhiza uralensis, a medicinal legume. Plant J. Cell Mol. Biol. 2017, 89, 181–194. [Google Scholar] [CrossRef]
- Nomura, Y.; Seki, H.; Suzuki, T.; Ohyama, K.; Mizutani, M.; Kaku, T.; Tamura, K.; Ono, E.; Horikawa, M.; Sudo, H.; et al. Functional specialization of UDP -glycosyltransferase 73P12 in licorice to produce a sweet triterpenoid saponin, glycyrrhizin. Plant J. 2019, 99, 1127–1143. [Google Scholar] [CrossRef] [Green Version]
- Udvardi, M.K.; Czechowski, T.; Scheible, W.-R. Eleven golden rules of quantitative RT-PCR. Plant Cell 2008, 20, 1736–1737. [Google Scholar] [CrossRef] [Green Version]
- Phillips, M.A.; D’Auria, J.C.; Luck, K.; Gershenzon, J. Evaluation of candidate reference genes for real-time quantitative PCR of plant samples using purified cDNA as template. Plant Mol. Biol. Report. 2009, 27, 407–416. [Google Scholar] [CrossRef] [Green Version]
- Hong, S.Y.; Seo, P.J.; Yang, M.; Xiang, F.; Park, C. Exploring valid reference genes for gene expression studies inBrachypodium distachyonby real-time PCR. BMC Plant Biol. 2008, 8, 112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dheda, K. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 2004, 37, 118–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radonić, A.; Thulke, S.; Mackay, I.M.; Landt, O.; Siegert, W.; Nitsche, A. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2004, 313, 856–862. [Google Scholar] [CrossRef]
- Nicot, N.; Hausman, J.F.; Hoffmann, L.; Evers, D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef]
- Le, D.T.; Nishiyama, R.; Watanabe, Y.; Tanaka, M.; Seki, M.; Ham, L.H.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.-S.P.; Shiu, S.-H. Differential Gene Expression in Soybean Leaf Tissues at Late Developmental Stages under Drought Stress Revealed by Genome-Wide Transcriptome Analysis. PLoS ONE 2012, 7, e49522. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Bao, J.D.; Dai, J.S.; Li, Y.; Zhu, Y. Genome-wide identification of new reference genes for qRT-PCR normalization under high temperature stress in rice endosperm. PLoS ONE 2015, 10, e0142015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bustin, S.A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Haraguchi, H. Antioxidative plants constituents. In Bioact. Compd. Nat. Sources; Tringali, C., Ed.; Taylor& Francis: London, UK, 2001; Chapter 9; pp. 337–378. [Google Scholar]
- Hayashi, H.; Sudo, H. Economic importance of licorice. Plant Biotechnol. 2009, 26, 101–104. [Google Scholar] [CrossRef] [Green Version]
- Hatano, T.; Shintani, Y.; Aga, Y.; Shiota, S.; Tsuchiya, T.; Yoshida, T. Phenolic constituents of licorice. VIII. Structures of glicophenone and glicoisoflavanone, and effects of licorice phenolics on methicillin-resistant Staphylococcus aureus. Chem. Pharm. Bull. 2000, 48, 1286–1292. [Google Scholar] [CrossRef] [Green Version]
- Xie, W.; Hao, Z.; Zhou, X.; Jiang, X.; Xu, L.; Wu, S.; Zhao, A.; Zhang, X.; Chen, B. Arbuscular mycorrhiza facilitates the accumulation of glycyrrhizin and liquiritin in Glycyrrhiza uralensis under drought stress. Mycorrhiza 2018, 28, 285–300. [Google Scholar] [CrossRef] [PubMed]
- Peleg, Z.; Blumwald, E. Hormone balance and abiotic stress tolerance in crop plants. Curr. Opin. Plant Biol. 2011, 14, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Geyter, N.; Gholami, A.; Goormachtig, S.; Goossens, A. Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci. 2012, 17, 349–359. [Google Scholar] [CrossRef]
- Maroufi, A. Selection of reference genes for real-time quantitative PCR analysis of gene expression in Glycyrrhiza glabra under drought stress. Biol. Plant. 2016, 60, 1–10. [Google Scholar] [CrossRef]
- Seki, H.; Ohyama, K.; Sawai, S.; Mizutani, M.; Ohnishi, T.; Sudo, H.; Akashi, T.; Aoki, T.; Saito, K.; Muranaka, T. Licorice β-amyrin 11-oxidase, a cytochrome P450 with a key role in the biosynthesis of the triterpene sweetener glycyrrhizin. Proc. Natl. Acad. Sci. USA 2008, 105, 14204–14209. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, H.; Huang, P.; Kirakosyan, A.; Inoue, K.; Hiraoka, N.; Ikeshiro, Y.; Kushiro, T.; Shibuya, M.; Ebizuka, Y. Cloning and characterization of a cDNA encoding beta-amyrin synthase involved in glycyrrhizin and soyasaponin biosyntheses in licorice. Biol. Pharm. Bull. 2001, 24, 912–916. [Google Scholar] [CrossRef] [Green Version]
- Nasrollahi, V.; Mirzaie-Asl, A.; Piri, K.; Nazeri, S.; Mehrabi, R. The effect of drought stress on the expression of key genes involved in the biosynthesis of triterpenoid saponins in liquorice (Glycyrrhiza glabra). Phytochemistry 2014, 103, 32–37. [Google Scholar] [CrossRef]
- Castro, P.; Roman, B.; Rubio, J.; Die, J.V. Selection of reference genes for expression studies in Cicer arietinum L.: Analysis of cyp81E3 gene expression against Ascochyta rabiei. Mol. Breed. 2012, 29, 261–274. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Abbas, M.; Wen, Y.; Niu, D.; Wang, L.; Sun, Y.; Li, Y. Selection and validation of reference genes for quantitative gene expression analyses in black locust (Robinia pseudoacacia L.) using real-time quantitative PCR. PLoS ONE 2018, 13, e0193076. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Liu, M.; Shi, J.; Zheng, G.; Wang, Y.; Wang, J.; Chen, Y.; Lu, C.; Yin, W. Reference gene selection for qPCR in Ammopiptanthus mongolicus under abiotic stresses and expression analysis of seven ROS-scavenging enzyme genes. Plant Cell Rep. 2012, 31, 1245–1254. [Google Scholar] [CrossRef]
- Barbosa Amorim, L.L.; Costa Ferreira-Neto, J.R.; Bezerra-Neto, J.P.; Pandolfi, V.; de Araujo, F.T.; da Silva Matos, M.K.; Santos, M.G.; Kido, E.A.; Benko-Iseppon, A.M. Cowpea and abiotic stresses: Identification of reference genes for transcriptional profiling by qPCR. Plant Methods 2018, 14. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Fu, Y.; Ban, L.; Wang, Z.; Feng, G.; Li, J.; Gao, H. Selection of reliable reference genes for quantitative real-time RT-PCR in alfalfa. Genes Genet. Syst. 2015, 90, 175–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kundu, A.; Patel, A.; Pal, A. Defining reference genes for qPCR normalization to study biotic and abiotic stress responses in Vigna mungo. Plant Cell Rep. 2013, 32, 1647–1658. [Google Scholar] [CrossRef]
- Jaiswal, P.S.; Kaur, N.; Randhawa, G.S. Identification of reference genes for qRT-PCR gene expression studies during seed development and under abiotic stresses in Cyamopsis tetragonoloba. Crop Sci. 2019, 59, 252–265. [Google Scholar] [CrossRef]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.-R. Genome-Wide Identification and Testing of Superior Reference Genes for Transcript Normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cordoba, E.M.; Die, J.V.; Gonzalez-Verdejo, C.I.; Nadal, S.; Roman, B. Selection of reference genes in Hedysarum coronarium under various stresses and stages of development. Anal. Biochem. 2011, 409, 236–243. [Google Scholar] [CrossRef]
- Zhang, Y.; Han, X.; Chen, S.; Zheng, L.; He, X.; Liu, M.; Qiao, G.; Wang, Y.; Zhuo, R. Selection of suitable reference genes for quantitative real-time PCR gene expression analysis in Salix matsudana under different abiotic stresses. Sci. Rep. 2017, 7, 40290. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Zhu, Q.; Li, J.; Yu, J.; Li, Y.; Huang, X.; Wang, W.; Tan, R.; Zhou, J.; Liao, H. Selection and evaluation of reference genes for expression analysis of Cassi. Biosci. Biotechnol. Biochem. 2015, 79, 1818–1826. [Google Scholar] [CrossRef] [Green Version]
- Ma, S.; Niu, H.; Liu, C.; Zhang, J.; Hou, C.; Wang, D. Expression stabilities of candidate reference genes for RT-qPCR under cifferent stress conditions in soybean. PLoS ONE 2013, 8, e75271. [Google Scholar] [CrossRef] [Green Version]
- Chi, X.; Hu, R.; Yang, Q.; Zhang, X.; Pan, L.; Chen, N.; Chen, M.; Yang, Z.; Wang, T.; He, Y.; et al. Validation of reference genes for gene expression studies in peanut by quantitative real-time RT-PCR. Mol. Genet. Genom. 2012, 287, 167–176. [Google Scholar] [CrossRef]
- Pereira, W.J.; Bassinello, P.Z.; Brondani, C.; Vianello, R.P. An improved method for RNA extraction from common bean seeds and validation of reference genes for qPCR. Crop Breed. Appl. Biotechnol. 2017, 17, 150–158. [Google Scholar] [CrossRef]
- Hayashi, H.; Huang, P.; Takada, S.; Obinata, M.; Inoue, K.; Shibuya, M.; Ebizuka, Y. Differential Expression of Three Oxidosqualene Cyclase mRNAs in Glycyrrhiza glabra. Biol. Pharm. Bull. 2004, 27, 1086–1092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, S.; Yongliang, L.; Min, W.; Xiaomin, L.; Xiaofei, S.; Chunzhao, L.; Ying, W.; Ji-Hong, L. Identification and validation of reference genes for quantitative teal-time PCR normalization and Its applications in Lycium. PLoS ONE 2014, 9, e97039. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR Data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Arocho, A.; Chen, B.; Ladanyi, M.; Pan, Q. Validation of the 2-DeltaDeltaCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagn Mol. Pathol. 2006, 15, 56–61. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef] [PubMed]








| Conditions | G. uralensis | G. inflata | ||
|---|---|---|---|---|
| Leaf | Root | Leaf | Root | |
| Control | CS, DNAJ, DREB | DNAJ, UBC2, CAC | COPS3, TUB, RAN | CS, CAC, R3HDM2 |
| Osmotic stress | TIF1, DNAJ, RAN | DNAJ, CAC, DREB | TUB, COPS3, DNAJ | TIF1, R3HDM2, ABCC2 |
| Salt stress | DNAJ, CYP, CAC | DREB, DNAJ, UBC2 | CAC, RAN, COPS3 | ABCC2, CAC, DNAJ |
| ABA treatment | CS, CAC, UBC2 | CAC, RAN, UBC2 | UBC2, CS, DREB | CAC, TUB, CS |
| MeJA treatment | ABCC2, UBC2, CAC | CAC, TUB, ABCC2 | RAN, COPS3, UBC2 | UBC2, DREB, CS |
| Gene | Description | Accession Number | Primer Sequence (5’–3’) Forward/Reverse | Amplicon Length (bp) | Tm (°C) | E (%) * |
|---|---|---|---|---|---|---|
| ACT | Actin1 | MW119712 | CCCACTCAACCCAAAGGC/TAACCCTCATAGATTGGCACAG | 183 | 62.8 | 92.72 |
| CAC | Clathrin complex AP1 | MW116276 | GAGTTTCAGCTTCCTCCTTGCA/TGATGGGGCTTTATCCTTTGG | 126 | 63.4 | 116.84 |
| CYP | Cyclophilin | MW119709 | AAGACGGAGTGGCTGGACG/TCTTGCCGGAGCTGGACC | 103 | 67 | 92.9 |
| DNAJ | Heat-shock protein 40 | MW116277 | TGGTTGTCAAGGAACTGGTATG/CACTGTGGGCAGCGGTCT | 135 | 63.4 | 91.94 |
| DREB | Dehydration responsive element binding | MW119710 | GGTTGCTGAAATTCGGGAGC/CATTGGGGAAGTTGAGGCG | 139 | 64 | 97.83 |
| EF1 | Translation elongation factor1 | MW116273 | GACTGGTACAAGGGACCAAC/AGACATCCTGCAATGGAAGC | 101 | 63.1 | 90.42 |
| RAN | Ras related protein | MW116274 | ACAGAGCAGACGATGACTACGA/CTGAGCCTTGATGACTTTGGA | 185 | 63.2 | 91.22 |
| TIF1 | Translation initiation factor | MW122063 | ACAACCGTTCAGGGATTGA/GGGTCCTGAACAACTGTACC | 98 | 62.2 | 77.95 |
| TUB | β-Tubulin | MW119713 | CCTTGAGCCAGGCACCAT/GTCCTTTCGCCCAGTTGTT | 113 | 63.6 | 86.97 |
| UBC2 | Ubiquitin-conjugating enzyme E2 | MW116271 | CTTCAACAAGACCCACCTGC/ACGTGCCTCCATCCCATG | 112 | 64.1 | 93.51 |
| ABCC2 | ATP binding-box transporter 2 | MW116275 | TGAGTCTTTCCAGGGCTTTATT/ATGGTGTTAAGGCGATGAGC | 160 | 62.7 | 90.63 |
| COPS3 | COP9 signal complex subunit 3 | MW119711 | GGAAGCGCCAATACGAGG/ACAACAAGCACAGCAGAAGAAA | 113 | 63.4 | 92.32 |
| CS | Citrate synthase | MW116272 | GCTCAGCCGTTGACCCAG/CACCACCAGGAAAAGCACC | 93 | 64.2 | 107.58 |
| R3HDM2 | R3H domain protein 2 | MW119714 | GCTTTGGGTTCAATGGAGG/TCAGCAGAGTGCTGGGGTC | 115 | 61.9 | 98.12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Liang, X.; Zhou, X.; Wu, Z.; Yuan, L.; Wang, Y.; Li, Y. Selection of Reference Genes for qRT-PCR Analysis in Medicinal Plant Glycyrrhiza under Abiotic Stresses and Hormonal Treatments. Plants 2020, 9, 1441. https://doi.org/10.3390/plants9111441
Li Y, Liang X, Zhou X, Wu Z, Yuan L, Wang Y, Li Y. Selection of Reference Genes for qRT-PCR Analysis in Medicinal Plant Glycyrrhiza under Abiotic Stresses and Hormonal Treatments. Plants. 2020; 9(11):1441. https://doi.org/10.3390/plants9111441
Chicago/Turabian StyleLi, Yuping, Xiaoju Liang, Xuguo Zhou, Zhigeng Wu, Ling Yuan, Ying Wang, and Yongqing Li. 2020. "Selection of Reference Genes for qRT-PCR Analysis in Medicinal Plant Glycyrrhiza under Abiotic Stresses and Hormonal Treatments" Plants 9, no. 11: 1441. https://doi.org/10.3390/plants9111441
APA StyleLi, Y., Liang, X., Zhou, X., Wu, Z., Yuan, L., Wang, Y., & Li, Y. (2020). Selection of Reference Genes for qRT-PCR Analysis in Medicinal Plant Glycyrrhiza under Abiotic Stresses and Hormonal Treatments. Plants, 9(11), 1441. https://doi.org/10.3390/plants9111441