CRISPR-Powered Microfluidics in Diagnostics: A Review of Main Applications
Abstract
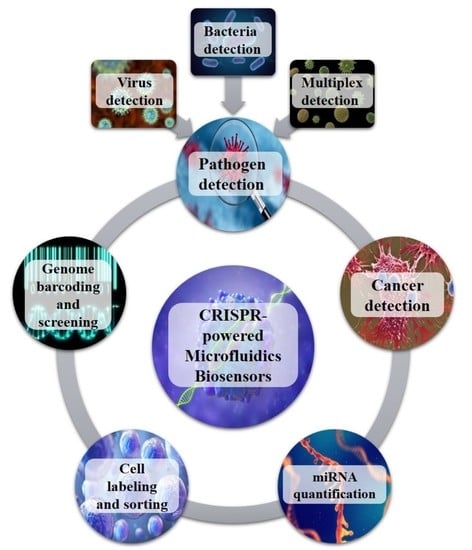
:1. Introduction
2. CRISPR-Powered Microfluidics Applications
2.1. Microfluidic CRISPR-Based Biosensors for Virus
2.1.1. SARS-CoV-2
2.1.2. Ebola Virus
2.1.3. Human Immunodeficiency (HIV)
2.1.4. Human Papillomavirus (HPV)
2.2. Microfluidic CRISPR-Based Biosensors for Bacteria Detection
2.3. Microfluidic CRISPR-Based Biosensors for Multiple Species
2.4. Microfluidic CRISPR-Based Biosensors for Single Nucleotide Polymorphisms (SNPs)
2.5. Microfluidic CRISPR-Based Biosensors for miRNA Quantification
2.6. Microfluidic CRISPR-Based Cell Labeling and Sorting Assay
2.7. Microfluidic CRISPR-Based in Genomics Studies
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cyranoski, D. Chinese scientists to pioneer first human CRISPR trial. Nature 2016, 535, 476–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brooks, A.K.; Gaj, T. Innovations in CRISPR technology. Curr. Opin. Biotechnol. 2018, 52, 95–101. [Google Scholar] [CrossRef]
- Adli, M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018, 9, 1911. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, Y.; Hong, W.; Huang, M.; Wu, M.; Zhao, X. Applications of genome editing technology in the targeted therapy of human diseases: Mechanisms, advances and prospects. Signal Transduct. Target. Ther. 2020, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Komaroff, A.L. Gene Editing Using CRISPR: Why the Excitement? JAMA 2017, 318, 699–700. [Google Scholar] [CrossRef] [PubMed]
- Han, H.A.; Pang, J.K.S.; Soh, B.-S. Mitigating off-target effects in CRISPR/Cas9-mediated in vivo gene editing. J. Mol. Med. 2020, 98, 615–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaj, T.; Gersbach, C.A.; Barbas III, C.F. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.; Chen, X.; Zhang, M.; Wang, X.; Chen, Y.; Qian, C.; Wu, J.; Xu, J. Versatile detection with CRISPR/Cas system from applications to challenges. TrAC Trends Anal. Chem. 2021, 135, 116150. [Google Scholar] [CrossRef]
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Mark, D.; Haeberle, S.; Roth, G.; von Stetten, F.; Zengerle, R. Microfluidic lab-on-a-chip platforms: Requirements, characteristics and applications. Chem. Soc. Rev. 2010, 39, 1153–1182. [Google Scholar] [CrossRef] [Green Version]
- Peyravian, N.; Malekzadeh Kebria, M.; Kiani, J.; Brouki Milan, P.; Mozafari, M. CRISPR-Associated (CAS) Effectors Delivery via Microfluidic Cell-Deformation Chip. Materials 2021, 14, 3164. [Google Scholar] [CrossRef] [PubMed]
- Melin, J.; Quake, S.R. Microfluidic Large-Scale Integration: The Evolution of Design Rules for Biological Automation. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 213–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buckner, R.L.; Krienen, F.M.; Castellanos, A.; Diaz, J.C.; Yeo, B.T.T. The organization of the human cerebellum estimated by intrinsic functional connectivity. J. Neurophysiol. 2011, 106, 2322–2345. [Google Scholar] [CrossRef] [PubMed]
- Sackmann, E.K.; Fulton, A.L.; Beebe, D.J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Balbino, T.A.; Azzoni, A.R.; de la Torre, L.G. Microfluidic devices for continuous production of pDNA/cationic liposome complexes for gene delivery and vaccine therapy. Colloids Surf. B Biointerfaces 2013, 111, 203–210. [Google Scholar] [CrossRef]
- Zhang, B. CRISPR/Cas gene therapy. J. Cell. Physiol. 2021, 236, 2459–2481. [Google Scholar] [CrossRef]
- Song, K.; Li, G.; Zu, X.; Du, Z.; Liu, L.; Hu, Z. The Fabrication and Application Mechanism of Microfluidic Systems for High Throughput Biomedical Screening: A Review. Micromachines 2020, 11, 297. [Google Scholar] [CrossRef] [Green Version]
- Haji Mohammadi, M.; Mulder, S.; Khashayar, P.; Kalbasi, A.; Azimzadeh, M.; Aref, A.R. Saliva Lab-on-a-chip biosensors: Recent novel ideas and applications in disease detection. Microchem. J. 2021, 168, 106506. [Google Scholar] [CrossRef]
- Halldorsson, S.; Lucumi, E.; Gómez-Sjöberg, R.; Fleming, R.M.T. Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices. Biosens. Bioelectron. 2015, 63, 218–231. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Shoaie, N.; Jahanpeyma, F.; Zhao, J.; Azimzadeh, M.; Al−Jamal, K.T. Optical, electrochemical and electrical (nano)biosensors for detection of exosomes: A comprehensive overview. Biosens. Bioelectron. 2020, 161, 112222. [Google Scholar] [CrossRef]
- Khodamoradi, M.; Rafizadeh Tafti, S.; Mousavi Shaegh, S.A.; Aflatoonian, B.; Azimzadeh, M.; Khashayar, P. Recent Microfluidic Innovations for Sperm Sorting. Chemosensors 2021, 9, 126. [Google Scholar] [CrossRef]
- Sinha, H.; Quach, A.B.V.; Vo, P.Q.N.; Shih, S.C.C. An automated microfluidic gene-editing platform for deciphering cancer genes. Lab Chip 2018, 18, 2300–2312. [Google Scholar] [CrossRef]
- Blondal, T.; Gamba, C.; Møller Jagd, L.; Su, L.; Demirov, D.; Guo, S.; Johnston, C.M.; Riising, E.M.; Wu, X.; Mikkelsen, M.J.; et al. Verification of CRISPR editing and finding transgenic inserts by Xdrop indirect sequence capture followed by short- and long-read sequencing. Methods 2021, 191, 68–77. [Google Scholar] [CrossRef]
- Park, J.S.; Hsieh, K.; Chen, L.; Kaushik, A.; Trick, A.Y.; Wang, T.H. Digital CRISPR/Cas-Assisted Assay for Rapid and Sensitive Detection of SARS-CoV-2. Adv. Sci. 2021, 8. [Google Scholar] [CrossRef] [PubMed]
- Bruch, R.; Johnston, M.; Kling, A.; Mattmüller, T.; Baaske, J.; Partel, S.; Madlener, S.; Weber, W.; Urban, G.A.; Dincer, C. CRISPR-powered electrochemical microfluidic multiplexed biosensor for target amplification-free miRNA diagnostics. Biosens. Bioelectron. 2021, 177. [Google Scholar] [CrossRef]
- Chen, Y.; Mei, Y.; Zhao, X.; Jiang, X. Reagents-Loaded, Automated Assay that Integrates Recombinase-Aided Amplification and Cas12a Nucleic Acid Detection for a Point-of-Care Test. Anal. Chem. 2020, 92, 14846–14852. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Mei, Y.; Jiang, X. Universal and high-fidelity DNA single nucleotide polymorphism detection based on a CRISPR/Cas12a biochip. Chem. Sci. 2021, 12, 4455–4462. [Google Scholar] [CrossRef]
- Phaneuf, C.R.; Seamon, K.J.; Eckles, T.P.; Sinha, A.; Schoeniger, J.S.; Harmon, B.; Meagher, R.J.; Abhyankar, V.V.; Koh, C.Y. Ultrasensitive multi-species detection of CRISPR-Cas9 by a portable centrifugal microfluidic platform. Anal. Methods 2019, 11, 559–565. [Google Scholar] [CrossRef]
- Chen, F.-E.; Lee, P.-W.; Trick, A.Y.; Park, J.S.; Chen, L.; Shah, K.; Mostafa, H.; Carroll, K.C.; Hsieh, K.; Wang, T.-H. Point-of-care CRISPR-Cas-assisted SARS-CoV-2 detection in an automated and portable droplet magnetofluidic device. Biosens. Bioelectron. 2021, 190, 113390. [Google Scholar] [CrossRef]
- Lee, H.; Choi, J.; Jeong, E.; Baek, S.; Kim, H.C.; Chae, J.H.; Koh, Y.; Seo, S.W.; Kim, J.S.; Kim, S.J. DCas9-mediated Nanoelectrokinetic Direct Detection of Target Gene for Liquid Biopsy. Nano Lett. 2018, 18, 7642–7650. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Feng, Q.; Han, Z.; Jiang, X. Enhancing gene editing efficiency for cells by CRISPR/Cas9 system-loaded multilayered nanoparticles assembled via microfluidics. Chin. J. Chem. Eng. 2021, 38, 216–220. [Google Scholar] [CrossRef]
- Silva, F.S.R.; Erdogmus, E.; Shokr, A.; Kandula, H.; Thirumalaraju, P.; Kanakasabapathy, M.K.; Hardie, J.M.; Pacheco, L.G.C.; Li, J.Z.; Kuritzkes, D.R.; et al. SARS-CoV-2 RNA Detection by a Cellphone-Based Amplification-Free System with CRISPR/CAS-Dependent Enzymatic (CASCADE) Assay. Adv. Mater. Technol. 2021. [Google Scholar] [CrossRef]
- Shokr, A.; Pacheco, L.G.C.; Thirumalaraju, P.; Kanakasabapathy, M.K.; Gandhi, J.; Kartik, D.; Silva, F.S.R.; Erdogmus, E.; Kandula, H.; Luo, S.; et al. Mobile Health (mHealth) Viral Diagnostics Enabled with Adaptive Adversarial Learning. ACS Nano 2021, 15, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, A.; Huyke, D.A.; Sharma, E.; Sahoo, M.K.; Huang, C.; Banaei, N.; Pinsky, B.A.; Santiago, J.G. Electric field-driven microfluidics for rapid CRISPR-based diagnostics and its application to detection of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 29518–29525. [Google Scholar] [CrossRef] [PubMed]
- de Puig, H.; Lee, R.A.; Najjar, D.; Tan, X.; Soeknsen, L.R.; Angenent-Mari, N.M.; Donghia, N.M.; Weckman, N.E.; Ory, A.; Ng, C.F.; et al. Minimally instrumented SHERLOCK (miSHERLOCK) for CRISPR-based point-of-care diagnosis of SARS-CoV-2 and emerging variants. Sci. Adv. 2021, 7. [Google Scholar] [CrossRef] [PubMed]
- Qin, P.; Park, M.; Alfson, K.J.; Tamhankar, M.; Carrion, R.; Patterson, J.L.; Griffiths, A.; He, Q.; Yildiz, A.; Mathies, R.; et al. Rapid and Fully Microfluidic Ebola Virus Detection with CRISPR-Cas13a. ACS Sens. 2019, 4, 1048–1054. [Google Scholar] [CrossRef]
- Gayet, R.V.; de Puig, H.; English, M.A.; Soenksen, L.R.; Nguyen, P.Q.; Mao, A.S.; Angenent-Mari, N.M.; Collins, J.J. Creating CRISPR-responsive smart materials for diagnostics and programmable cargo release. Nat. Protoc. 2020, 15, 3030–3063. [Google Scholar] [CrossRef]
- Yin, K.; Ding, X.; Li, Z.; Zhao, H.; Cooper, K.; Liu, C. Dynamic Aqueous Multiphase Reaction System for One-Pot CRISPR-Cas12a-Based Ultrasensitive and Quantitative Molecular Diagnosis. Anal. Chem. 2020, 92, 8561–8568. [Google Scholar] [CrossRef]
- Ackerman, C.M.; Myhrvold, C.; Thakku, S.G.; Freije, C.A.; Metsky, H.C.; Yang, D.K.; Ye, S.H.; Boehm, C.K.; Kosoko-Thoroddsen, T.S.F.; Kehe, J.; et al. Massively multiplexed nucleic acid detection with Cas13. Nature 2020, 582, 277–282. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef] [Green Version]
- Bruch, R.; Baaske, J.; Chatelle, C.; Meirich, M.; Madlener, S.; Weber, W.; Dincer, C.; Urban, G.A. CRISPR/Cas13a-Powered Electrochemical Microfluidic Biosensor for Nucleic Acid Amplification-Free miRNA Diagnostics. Adv. Mater. 2019, 31, 1905311. [Google Scholar] [CrossRef] [Green Version]
- Chung, Y.-S.; Lee, N.-J.; Woo, S.H.; Kim, J.-M.; Kim, H.M.; Jo, H.J.; Park, Y.E.; Han, M.-G. Validation of real-time RT-PCR for detection of SARS-CoV-2 in the early stages of the COVID-19 outbreak in the Republic of Korea. Sci. Rep. 2021, 11, 14817. [Google Scholar] [CrossRef] [PubMed]
- Ganbaatar, U.; Liu, C. CRISPR-Based COVID-19 Testing: Toward Next-Generation Point-of-Care Diagnostics. Front. Cell. Infect. Microbiol. 2021, 11, 663949. [Google Scholar] [CrossRef] [PubMed]
- Di Paola, N.; Sanchez-Lockhart, M.; Zeng, X.; Kuhn, J.H.; Palacios, G. Viral genomics in Ebola virus research. Nat. Rev. Microbiol. 2020, 18, 365–378. [Google Scholar] [CrossRef]
- McLaren, P.J.; Fellay, J. HIV-1 and human genetic variation. Nat. Rev. Genet. 2021, 22, 645–657. [Google Scholar] [CrossRef] [PubMed]
- Bordigoni, A.; Motte, A.; Tissot-Dupont, H.; Colson, P.; Desnues, C. Development and validation of a multiplex qPCR assay for detection and relative quantification of HPV16 and HPV18 E6 and E7 oncogenes. Sci. Rep. 2021, 11, 4039. [Google Scholar] [CrossRef] [PubMed]
- Hattakam, S.; Elong Ngono, A.; McCauley, M.; Shresta, S.; Yamabhai, M. Repeated exposure to dengue virus elicits robust cross neutralizing antibodies against Zika virus in residents of Northeastern Thailand. Sci. Rep. 2021, 11, 9634. [Google Scholar] [CrossRef]
- Labib, M.; Philpott, D.N.; Wang, Z.; Nemr, C.; Chen, J.B.; Sargent, E.H.; Kelley, S.O. Magnetic Ranking Cytometry: Profiling Rare Cells at the Single-Cell Level. Acc. Chem. Res. 2020, 53, 1445–1457. [Google Scholar] [CrossRef]
- Wang, Z.; Gagliardi, M.; Mohamadi, R.M.; Ahmed, S.U.; Labib, M.; Zhang, L.; Popescu, S.; Zhou, Y.; Sargent, E.H.; Keller, G.M.; et al. Ultrasensitive and rapid quantification of rare tumorigenic stem cells in hPSC-derived cardiomyocyte populations. Sci. Adv. 2020, 6, eaay7629. [Google Scholar] [CrossRef] [Green Version]
- Aldridge, P.M.; Mukhopadhyay, M.; Ahmed, S.U.; Zhou, W.; Christinck, E.; Makonnen, R.; Sargent, E.H.; Kelley, S.O. Prismatic Deflection of Live Tumor Cells and Cell Clusters. ACS Nano 2018, 12, 12692–12700. [Google Scholar] [CrossRef]
- Mair, B.; Aldridge, P.M.; Atwal, R.S.; Philpott, D.; Zhang, M.; Masud, S.N.; Labib, M.; Tong, A.H.Y.; Sargent, E.H.; Angers, S.; et al. High-throughput genome-wide phenotypic screening via immunomagnetic cell sorting. Nat. Biomed. Eng. 2019, 3, 796–805. [Google Scholar] [CrossRef]
- Parvez, S.; Herdman, C.; Beerens, M.; Chakraborti, K.; Harmer, Z.P.; Yeh, J.-R.J.; MacRae, C.A.; Yost, H.J.; Peterson, R.T. MIC-Drop: A platform for large-scale in vivo CRISPR screens. Science 2021, 373, 1146–1151. [Google Scholar] [CrossRef]
- Elowitz, M.B.; Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 2000, 403, 335–338. [Google Scholar] [CrossRef] [PubMed]
- Henningsen, J.; Schwarz-Schilling, M.; Leibl, A.; Gutiérrez, J.; Sagredo, S.; Simmel, F.C. Single Cell Characterization of a Synthetic Bacterial Clock with a Hybrid Feedback Loop Containing dCas9-sgRNA. ACS Synth. Biol. 2020, 9, 3377–3387. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Jendresen, C.B.; Grünberger, A.; Ronda, C.; Jensen, S.I.; Noack, S.; Nielsen, A.T. Enhanced protein and biochemical production using CRISPRi-based growth switches. Metab. Eng. 2016, 38, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Camsund, D.; Lawson, M.J.; Larsson, J.; Jones, D.; Zikrin, S.; Fange, D.; Elf, J. Time-resolved imaging-based CRISPRi screening. Nat. Methods 2020, 17, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Kong, T.; Backes, N.; Kalwa, U.; Legner, C.; Phillips, G.J.; Pandey, S. Adhesive Tape Microfluidics with an Autofocusing Module That Incorporates CRISPR Interference: Applications to Long-Term Bacterial Antibiotic Studies. ACS Sens. 2019, 4, 2638–2645. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Liu, Z.; Zhao, L.; Wang, F.; Yu, Y.; Yang, J.; Chen, R.; Qin, L. Microfluidic Cell Deformability Assay for Rapid and Efficient Kinase Screening with the CRISPR-Cas9 System. Angew. Chem. Int. Ed. 2016, 55, 8561–8565. [Google Scholar] [CrossRef]







| Analyte/Biomarker | Components, Mechanism, and Special Features | Sensitivity | References |
|---|---|---|---|
| RNA/DNA hybrid of SARS-CoV-2 genome | CRISPR/Cas12a, trans cleavage, catalase-ssDNA probe, no amplification step, magnetic beads, bubble image detection, visible readout, detection time: 71 min | 50 copies per µL | [32] |
| Intact virus and RNA detection of SARS-CoV-2 and HIV | Magnetic beads, platinum nanoparticles, antibodies, CRISPR/dCas9, RNA extraction and amplification, time: more than 60 min | SARS-CoV-2: 103 copies per mL, HIV: 250 copies per mL | [33] |
| SARS-CoV-2 genome and heat-inactivated SARS-CoV-2 virus | RT-RPA, CRISPR/Cas12a, Alexa647 fluorophore reporter, time of RNA detection: 15 min, time of inactivated virus detection: 30 min | RNA detection: 1 GE/µL; heat-inactivated detection: 20 GE/µL | [24] |
| SARS-CoV-2 genome | RT-RPA, CRISPR/Cas12a, magnetic beads, digital magnetofluidics device, fluorescent detectors, time: 30 min | 1 GE per µL | [29] |
| SARS-CoV-2 genome | RT-LAMP, CRISPR/Cas12, electrokinetic microfluidics, isotachophoresis, ion mobility, trans cleavage, time: 30–40 min | 10 copies per µL | [34] |
| SARS-CoV-2 genome | Amplification, CRISPR/Cas13, two-part miniaturized device, mobile software, quantify fluorescent readout, time: 55 min | 1200 copies per mL | [35] |
| Ebola genome | CRISPR/Cas13a, trans cleavage, 24 parallel assays, no amplification, time: 5 min | 20 PFU per mL, 5.45 × 107 copies per mL | [36] |
| Ebola genome | CRISPR/Cas12a, RT-RPA, trans cleavage, PEG-ssDNA, PA-DNA, µPAD, electronic detection, AuNPs, time: within a few hours | 11 aM nucleic acid | [37] |
| CCR5 gene, HIV model | CRISPR/dCas9, ion concentration polarization, ion mobility behavior, no amplification, time: 100 min | 3 pM DNA | [30] |
| HPV16 and HPV18 genome | CRISPR/Cas12a, dynamic aqueous multiphase detection, three-chambered microfluidic, RPA amplification, time: 60 min | HPV16: 10 copies per mL, HPV18: 100 copies per mL | [38] |
| Pseudomonas aeruginosa genome | CRISPR/Cas12a, recombinase-aided amplification, automated reaction, time: 90 min | 103 CFU per mL, 10 aM nucleic acid | [26] |
| Genomes of 169 viruses | CRISPR/Cas13, PCR or RPA amplification, trans cleavage, microarray chips, multiplex detection, time: 30–60 min | 104 copies per µL | [39] |
| Zika virus (ZIKV) and Flavivirus Dengue (DENV) genome | RPA amplification, CRISPR/Cas13, trans cleavage, lateral flow test, gold nanoparticles, FAM reporter, time: 90 min | 2 aM RNA | [40] |
| CYP1A1*2 gene (A4889G, rs1048943) | PAM insertion by PCR amplification, centrifugal microfluidic chip, simultaneous rotation, and fluorescent readout with point-of-care analytical instrument | three genotypes of target SNPs | [27] |
| miR-19b (brain tumor marker) | Amplification-free, CRISPR/Cas13a, microfluidic sensor, amperometry readout | 10 pM LOD | [41] |
| miR-19b and miRNA-20a | Amplification-free, CRISPR/Cas13a, microfluidic sensor, simultaneous multiplexed detection, amperometry | up to 8 miRNA simultaneously | [25] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azimzadeh, M.; Mousazadeh, M.; Jahangiri-Manesh, A.; Khashayar, P.; Khashayar, P. CRISPR-Powered Microfluidics in Diagnostics: A Review of Main Applications. Chemosensors 2022, 10, 3. https://doi.org/10.3390/chemosensors10010003
Azimzadeh M, Mousazadeh M, Jahangiri-Manesh A, Khashayar P, Khashayar P. CRISPR-Powered Microfluidics in Diagnostics: A Review of Main Applications. Chemosensors. 2022; 10(1):3. https://doi.org/10.3390/chemosensors10010003
Chicago/Turabian StyleAzimzadeh, Mostafa, Marziyeh Mousazadeh, Atieh Jahangiri-Manesh, Pouria Khashayar, and Patricia Khashayar. 2022. "CRISPR-Powered Microfluidics in Diagnostics: A Review of Main Applications" Chemosensors 10, no. 1: 3. https://doi.org/10.3390/chemosensors10010003
APA StyleAzimzadeh, M., Mousazadeh, M., Jahangiri-Manesh, A., Khashayar, P., & Khashayar, P. (2022). CRISPR-Powered Microfluidics in Diagnostics: A Review of Main Applications. Chemosensors, 10(1), 3. https://doi.org/10.3390/chemosensors10010003