Plasmonic Nanomaterials for Colorimetric Biosensing: A Review
Abstract
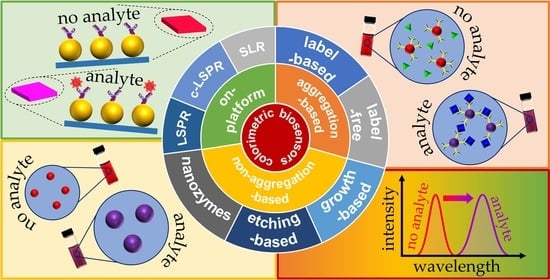
:1. Introduction
2. Platform-Based Colorimetric Biosensors
2.1. LSPR-Based Biosensing


2.2. c-LSPR-Based On-Platform Colorimetric Biosensors


2.3. SLR-Based On-Platform Colorimetric Biosensors


3. Nanoparticle Aggregation-Based Colorimetric Biosensing
3.1. Label-Based Colorimetric Biosensors


3.2. Label-Free Colorimetric Biosensors
4. Non-Aggregation-Based Colorimetric Biosensors
4.1. Nanozyme-Based Colorimetric Biosensors
4.2. Etching-Based Colorimetric Biosensors
4.3. Growth-Based Colorimetric Biosensors

5. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fang, Y.; Sun, M. Nanoplasmonic waveguides: Towards applications in integrated nanophotonic circuits. Light Sci. Appl. 2015, 4, e294. [Google Scholar] [CrossRef] [Green Version]
- El-Sayed, I.H.; Huang, X.; El-Sayed, M.A. Selective laser photo-thermal therapy of epithelial carcinoma using anti-EGFR antibody conjugated gold nanoparticles. Cancer Lett. 2006, 239, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Ferry, V.E.; Sweatlock, L.A.; Pacifici, D.; Atwater, H.A. Plasmonic nanostructure design for efficient light coupling into solar cells. Nano Lett. 2008, 8, 4391–4397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anker, J.N.; Hall, W.P.; Lyandres, O.; Shah, N.C.; Zhao, J.; Van Duyne, R.P. Biosensing with plasmonic nanosensors. Nat. Mater. 2008, 7, 442–453. [Google Scholar] [CrossRef] [PubMed]
- Koller, D.M.; Hohenau, A.; Ditlbacher, H.; Galler, N.; Reil, F.; Aussenegg, F.R.; Leitner, A.; List, E.J.W.; Krenn, J.R. Organic plasmon-emitting diode. Nat. Photonics 2008, 2, 684–687. [Google Scholar] [CrossRef]
- Akimov, A.V.; Mukherjee, A.; Yu, C.L.; Chang, D.E.; Zibrov, A.S.; Hemmer, P.R.; Park, H.; Lukin, M.D. Generation of single optical plasmons in metallic nanowires coupled to quantum dots. Nature 2007, 450, 402–406. [Google Scholar] [CrossRef] [PubMed]
- Pelton, M.; Aizpurua, J.; Bryant, G. Metal-nanoparticle plasmonics. Laser Photonics Rev. 2008, 2, 136–159. [Google Scholar] [CrossRef] [Green Version]
- Cao, X.; Wang, Z.; Bi, L.; Bi, C.; Du, Q. Gold nanocage-based surface-enhanced Raman scattering probes for long-term monitoring of intracellular microRNA during bone marrow stem cell differentiation. Nanoscale 2020, 12, 1513–1527. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xue, J.; Bi, C.; Xin, H.; Wang, Y.; Cao, X. Quantitative and specific detection of cancer-related microRNAs in living cells using surface-enhanced Raman scattering imaging based on hairpin DNA-functionalized gold nanocages. Analyst 2019, 144, 7250–7262. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.Y.; Yang, W.G.; Hong, S.H.; Kim, G.H.; Hwang, K.; Chae, W.S. Amplified fluorescence imaging using photonic Ag nanotip array: A comparative study on surface morphology effects. Appl. Surf. Sci. 2020, 529, 147139. [Google Scholar] [CrossRef]
- Fothergill, S.M.; Joyce, C.; Xie, F. Metal enhanced fluorescence biosensing: From ultra-violet towards second near-infrared window. Nanoscale 2018, 10, 20914–20929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minopoli, A.; Acunzo, A.; Della Ventura, B.; Velotta, R. Nanostructured Surfaces as Plasmonic Biosensors: A Review. Adv. Mater. Interfaces 2021, 2101133, 2101133. [Google Scholar] [CrossRef]
- Minopoli, A.; Della Ventura, B.; Campanile, R.; Tanner, J.A.; Offenhäusser, A.; Mayer, D.; Velotta, R. Randomly positioned gold nanoparticles as fluorescence enhancers in apta-immunosensor for malaria test. Microchim. Acta 2021, 188, 88. [Google Scholar] [CrossRef] [PubMed]
- Minopoli, A.; Scardapane, E.; Ventura, B.D.; Tanner, J.A.; Offenhäusser, A.; Mayer, D.; Velotta, R. Double-Resonant Nanostructured Gold Surface for Multiplexed Detection. ACS Appl. Mater. Interfaces 2022, 14, 6417–6427. [Google Scholar] [CrossRef]
- Pashchenko, O.; Shelby, T.; Banerjee, T.; Santra, S. A Comparison of Optical, Electrochemical, Magnetic, and Colorimetric Point-of-Care Biosensors for Infectious Disease Diagnosis. ACS Infect. Dis. 2018, 4, 1162–1178. [Google Scholar] [CrossRef] [PubMed]
- Aldewachi, H.; Chalati, T.; Woodroofe, M.N.; Bricklebank, N.; Sharrack, B.; Gardiner, P. Gold nanoparticle-based colorimetric biosensors. Nanoscale 2018, 10, 18–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, Y.; Hwang, J.H.; Lee, S.Y. Recent Trends in Nanomaterials-Based Colorimetric Detection of Pathogenic Bacteria and Viruses. Small Methods 2018, 2, 1700351. [Google Scholar] [CrossRef]
- Zhao, V.X.T.; Wong, T.I.; Zheng, X.T.; Tan, Y.N.; Zhou, X. Colorimetric biosensors for point-of-care virus detections. Mater. Sci. Energy Technol. 2020, 3, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Liu, B.; Wei, G. Two-dimensional material-based colorimetric biosensors: A review. Biosensors 2021, 11, 259. [Google Scholar] [CrossRef] [PubMed]
- Iravani, S. Nano-And biosensors for the detection of SARS-CoV-2: Challenges and opportunities. Mater. Adv. 2020, 1, 3092–3103. [Google Scholar] [CrossRef]
- Vázquez, M.; Anfossi, L.; Ben-Yoav, H.; Diéguez, L.; Karopka, T.; Della Ventura, B.; Abalde-Cela, S.; Minopoli, A.; Di Nardo, F.; Shukla, V.K.; et al. Use of some cost-effective technologies for a routine clinical pathology laboratory. Lab Chip 2021, 21, 4330–4351. [Google Scholar] [CrossRef] [PubMed]
- Olofsson, S.; Brittain-Long, R.; Andersson, L.M.; Westin, J.; Lindh, M. PCR for detection of respiratory viruses: Seasonal variations of virus infections. Expert Rev. Anti. Infect. Ther. 2011, 9, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Appak, Ö.; Duman, M.; Belet, N.; Sayiner, A.A. Viral respiratory infections diagnosed by multiplex polymerase chain reaction in pediatric patients. J. Med. Virol. 2019, 91, 731–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Yi, Y.; Luo, X.; Xiong, N.; Liu, Y.; Li, S.; Sun, R.; Wang, Y.; Hu, B.; Chen, W.; et al. Development and Clinical Application of A Rapid IgM-IgG Combined Antibody Test for SARS-CoV-2 Infection Diagnosis. J. Med. Virol. 2020, 92, 1518–1524. [Google Scholar] [CrossRef] [PubMed]
- Dou, M.; Zhu, K.; Fan, Z.; Zhang, Y.; Chen, X.; Zhou, X.; Ding, X.; Li, L.; Gu, Z.; Guo, M.; et al. Reproductive Hormones and Their Receptors May Affect Lung Cancer. Cell. Physiol. Biochem. 2017, 44, 1425–1434. [Google Scholar] [CrossRef] [PubMed]
- Maddali, H.; Miles, C.E.; Kohn, J.; O’Carroll, D.M. Optical Biosensors for Virus Detection: Prospects for SARS-CoV-2/COVID-19. ChemBioChem 2021, 22, 1176–1189. [Google Scholar] [CrossRef] [PubMed]
- Maier, S.A. Plasmonics: Fundamentals and Applications; Springer: New York, NY, USA, 2007; ISBN 0387331506. [Google Scholar]
- Mayer, K.M.; Hafner, J.H. Localized surface plasmon resonance sensors. Chem. Rev. 2011, 111, 3828–3857. [Google Scholar] [CrossRef] [PubMed]
- Kravets, V.G.; Kabashin, A.V.; Barnes, W.L.; Grigorenko, A.N. Plasmonic Surface Lattice Resonances: A Review of Properties and Applications. Chem. Rev. 2018, 118, 5912–5951. [Google Scholar] [CrossRef]
- Njoki, P.N.; Lim, I.I.S.; Mott, D.; Park, H.Y.; Khan, B.; Mishra, S.; Sujakumar, R.; Luo, J.; Zhong, C.J. Size correlation of optical and spectroscopic properties for gold nanoparticles. J. Phys. Chem. C 2007, 111, 14664–14669. [Google Scholar] [CrossRef]
- Ghosh, S.K.; Pal, T. Interparticle Coupling Effect on the Surface Plasmon Resonance of Gold Nanoparticles: From Theory to Applications. Chem. Rev. 2007, 107, 4797–4862. [Google Scholar] [CrossRef]
- Kaye, S.; Zeng, Z.; Sanders, M.; Chittur, K.; Koelle, P.M.; Lindquist, R.; Manne, U.; Lin, Y.; Wei, J. Label-free detection of DNA hybridization with a compact LSPR-based fiber-optic sensor. Analyst 2017, 142, 1974–1981. [Google Scholar] [CrossRef]
- Kawasaki, D.; Yamada, H.; Maeno, K.; Sueyoshi, K.; Hisamoto, H.; Endo, T. Core-Shell-Structured Gold Nanocone Array for Label-Free DNA Sensing. ACS Appl. Nano Mater. 2019, 2, 4983–4990. [Google Scholar] [CrossRef]
- Song, C.; Ding, W.; Zhao, W.; Liu, H.; Wang, J.; Yao, Y.; Yao, C. High peroxidase-like activity realized by facile synthesis of FeS2 nanoparticles for sensitive colorimetric detection of H2O2 and glutathione. Biosens. Bioelectron. 2020, 151, 111983. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhao, X.; Xu, Z. Plasmonic colorimetric biosensor for visual detection of telomerase activity based on horseradish peroxidase-encapsulated liposomes and etching of Au nanobipyramids. Sens. Actuators B Chem. 2019, 296, 126646. [Google Scholar] [CrossRef]
- Klimov, V. Nanoplasmonics; CRC Press: Boca Raton, FL, USA, 2013; ISBN 9789814267427. [Google Scholar]
- Karimi, S.; Moshaii, A.; Abbasian, S.; Nikkhah, M. Surface Plasmon Resonance in Small Gold Nanoparticles: Introducing a Size-Dependent Plasma Frequency for Nanoparticles in Quantum Regime. Plasmonics 2019, 14, 851–860. [Google Scholar] [CrossRef]
- Link, S.; El-Sayed, M.A. Size and temperature dependence of the plasmon absorption of colloidal gold nanoparticles. J. Phys. Chem. B 1999, 103, 4212–4217. [Google Scholar] [CrossRef]
- Jain, P.K.; Lee, K.S.; El-Sayed, I.H.; El-Sayed, M.A. Calculated absorption and scattering properties of gold nanoparticles of different size, shape, and composition: Applications in biological imaging and biomedicine. J. Phys. Chem. B 2006, 110, 7238–7248. [Google Scholar] [CrossRef] [Green Version]
- Shafiqa, A.R.; Abdul Aziz, A.; Mehrdel, B. Nanoparticle Optical Properties: Size Dependence of a Single Gold Spherical Nanoparticle. J. Phys. Conf. Ser. 2018, 1083, 012040. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.M.; Lazarides, A.A. Sensitivity of metal nanoparticle surface plasmon resonance to the dielectric environment. J. Phys. Chem. B 2005, 109, 21556–21565. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yang, J.; He, X.; Zhang, J.; Huang, J.; Chen, D.; Han, Y. Plasmonic refractive index sensor with high figure of merit based on concentric-rings resonator. Sensors 2018, 18, 116. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Xia, Y. Increased sensitivity of surface plasmon resonance of gold nanoshells compared to that of gold solid colloids in response to environmental changes. Anal. Chem. 2002, 74, 5297–5305. [Google Scholar] [CrossRef] [PubMed]
- Mock, J.J.; Smith, D.R.; Schultz, S. Local refractive index dependence of plasmon resonance spectra from individual nanoparticles. Nano Lett. 2003, 3, 485–491. [Google Scholar] [CrossRef]
- Chen, H.; Kou, X.; Yang, Z.; Ni, W.; Wang, J. Shape- and size-dependent refractive index sensitivity of gold nanoparticles. Langmuir 2008, 24, 5233–5237. [Google Scholar] [CrossRef] [PubMed]
- Sherry, L.J.; Chang, S.H.; Schatz, G.C.; Van Duyne, R.P.; Wiley, B.J.; Xia, Y. Localized surface plasmon resonance spectroscopy of single silver nanocubes. Nano Lett. 2005, 5, 2034–2038. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, A.; Khan, S.; Gupta, A.; Nandi, C.K. Effect of surface chemistry and morphology of gold nanoparticle on the structure and activity of common blood proteins. New J. Chem. 2016, 40, 4879–4883. [Google Scholar] [CrossRef]
- Austin Suthanthiraraj, P.P.; Sen, A.K. Localized surface plasmon resonance (LSPR) biosensor based on thermally annealed silver nanostructures with on-chip blood-plasma separation for the detection of dengue non-structural protein NS1 antigen. Biosens. Bioelectron. 2019, 132, 38–46. [Google Scholar] [CrossRef]
- Park, Y.; Ryu, B.; Deng, Q.; Pan, B.; Song, Y.; Tian, Y.; Alam, H.B.; Li, Y.; Liang, X.; Kurabayashi, K. An Integrated Plasmo-Photoelectronic Nanostructure Biosensor Detects an Infection Biomarker Accompanying Cell Death in Neutrophils. Small 2020, 16, 1905611. [Google Scholar] [CrossRef] [PubMed]
- Focsan, M.; Craciun, A.M.; Potara, M.; Leordean, C.; Vulpoi, A.; Maniu, D.; Astilean, S. Flexible and Tunable 3D Gold Nanocups Platform as Plasmonic Biosensor for Specific Dual LSPR-SERS Immuno-Detection. Sci. Rep. 2017, 7, 14240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badshah, M.A.; Koh, N.Y.; Zia, A.W.; Abbas, N.; Zahra, Z.; Saleem, M.W. Recent developments in plasmonic nanostructures for metal enhanced fluorescence-based biosensing. Nanomaterials 2020, 10, 1749. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.S.; El-Sayed, M.A. Gold and silver nanoparticles in sensing and imaging: Sensitivity of plasmon response to size, shape, and metal composition. J. Phys. Chem. B 2006, 110, 19220–19225. [Google Scholar] [CrossRef] [PubMed]
- Kazuma, E.; Tatsuma, T. Localized surface plasmon resonance sensors based on wavelength-tunable spectral dips. Nanoscale 2014, 6, 2397–2405. [Google Scholar] [CrossRef] [Green Version]
- Na, H.K.; Wi, J.S.; Son, H.Y.; Ok, J.G.; Huh, Y.M.; Lee, T.G. Discrimination of single nucleotide mismatches using a scalable, flexible, and transparent three-dimensional nanostructure-based plasmonic miRNA sensor with high sensitivity. Biosens. Bioelectron. 2018, 113, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Lednický, T.; Bonyár, A. Large Scale Fabrication of Ordered Gold Nanoparticle–Epoxy Surface Nanocomposites and Their Application as Label-Free Plasmonic DNA Biosensors. ACS Appl. Mater. Interfaces 2020, 12, 4804–4814. [Google Scholar] [CrossRef] [PubMed]
- Mock, J.J.; Barbic, M.; Smith, D.R.; Schultz, D.A.; Schultz, S. Shape effects in plasmon resonance of individual colloidal silver nanoparticles. J. Chem. Phys. 2002, 116, 6755–6759. [Google Scholar] [CrossRef]
- Wang, B.; Singh, S.C.; Lu, H.; Guo, C. Design of Aluminum Bowtie Nanoantenna Array with Geometrical Control to Tune LSPR from UV to Near-IR for Optical Sensing. Plasmonics 2020, 15, 609–621. [Google Scholar] [CrossRef]
- Qi, X.; Bi, J. Plasmonic sensors relying on nanoparticle arrays created by a template-directed dewetting process. Opt. Commun. 2019, 453, 124328. [Google Scholar] [CrossRef]
- Rapisarda, A.; Giamblanco, N.; Marletta, G. Kinetic discrimination of DNA single-base mutations by localized surface plasmon resonance. J. Colloid Interface Sci. 2017, 487, 141–148. [Google Scholar] [CrossRef]
- Zhen, Y.R.; Fung, K.H.; Chan, C.T. Collective plasmonic modes in two-dimensional periodic arrays of metal nanoparticles. Phys. Rev. B Condens. Matter Mater. Phys. 2008, 78, 035419. [Google Scholar] [CrossRef] [Green Version]
- Nordlander, P.; Oubre, C.; Prodan, E.; Li, K.; Stockman, M.I. Plasmon Hybridization in Nanoparticle Dimers. Nano Lett. 2004, 4, 899–903. [Google Scholar] [CrossRef]
- Prodan, E.; Nordlander, P. Plasmon hybridization in spherical nanoparticles. J. Chem. Phys. 2004, 120, 5444–5454. [Google Scholar] [CrossRef] [PubMed]
- Sheikholeslami, S.; Jun, Y.W.; Jain, P.K.; Alivisatos, A.P. Coupling of optical resonances in a compositionally asymmetric plasmonic nanoparticle dimer. Nano Lett. 2010, 10, 2655–2660. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.K.; Eustis, S.; El-Sayed, M.A. Plasmon Coupling in Nanorod Assemblies: Optical Absorption, Discrete Dipole Approximation Simulation, and Exciton-Coupling Model. J. Phys. Chem. B 2006, 110, 18243–18253. [Google Scholar] [CrossRef] [PubMed]
- Deng, T.-S.; Parker, J.; Yifat, Y.; Shepherd, N.; Scherer, N.F. Dark Plasmon Modes in Symmetric Gold Nanoparticle Dimers Illuminated by Focused Cylindrical Vector Beams. J. Phys. Chem. C 2018, 122, 27662–27672. [Google Scholar] [CrossRef]
- Jain, P.K.; Huang, W.; El-Sayed, M.A. On the Universal Scaling Behavior of the Distance Decay of Plasmon Coupling in Metal Nanoparticle Pairs: A Plasmon Ruler Equation. Nano Lett. 2007, 7, 2080–2088. [Google Scholar] [CrossRef]
- Jain, P.K.; El-Sayed, M.A. Plasmonic coupling in noble metal nanostructures. Chem. Phys. Lett. 2010, 487, 153–164. [Google Scholar] [CrossRef]
- Sadeghi, S.M.; Gutha, R.R. Coherent Networks of Plasmonic Dipole Domains: Long-Range Optical Coupling of Phase-Correlated Packages of Metallic Nanoparticles. Phys. Rev. Appl. 2021, 15, 034018. [Google Scholar] [CrossRef]
- Das, A.; Kumar, K.; Dhawan, A. Periodic arrays of plasmonic crossed-bowtie nanostructures interspaced with plasmonic nanocrosses for highly sensitive LSPR based chemical and biological sensing. RSC Adv. 2021, 11, 8096–8106. [Google Scholar] [CrossRef]
- Verellen, N.; Van Dorpe, P.; Huang, C.; Lodewijks, K.; Vandenbosch, G.A.E.; Lagae, L.; Moshchalkov, V.V. Plasmon line shaping using nanocrosses for high sensitivity localized surface plasmon resonance sensing. Nano Lett. 2011, 11, 391–397. [Google Scholar] [CrossRef]
- Zhou, L.; Ding, F.; Chen, H.; Ding, W.; Zhang, W.; Chou, S.Y. Enhancement of immunoassay’s fluorescence and detection sensitivity using three-dimensional plasmonic nano-antenna-dots array. Anal. Chem. 2012, 84, 4489–4495. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.C.; Chen, H.L. Nanoimprint technology for patterning functional materials and its applications. Microelectron. Eng. 2015, 132, 98–119. [Google Scholar] [CrossRef]
- Su, H.; Cheng, X.R.; Endo, T.; Kerman, K. Photonic crystals on copolymer film for label-free detection of DNA hybridization. Biosens. Bioelectron. 2018, 103, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Dickreuter, S.; Gleixner, J.; Kolloch, A.; Boneberg, J.; Scheer, E.; Leiderer, P. Mapping of plasmonic resonances in nanotriangles. Beilstein J. Nanotechnol. 2013, 4, 588–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, Y.; Lu, Y.; Gates, B.; Xia, Y. Template-assisted self-assembly: A practical route to complex aggregates of monodispersed colloids with well-defined sizes, shapes, and structures. J. Am. Chem. Soc. 2001, 123, 8718–8729. [Google Scholar] [CrossRef] [PubMed]
- Cheung, C.L.; Nikolić, R.J.; Reinhardt, C.E.; Wang, T.F. Fabrication of nanopillars by nanosphere lithography. Nanotechnology 2006, 17, 1339–1343. [Google Scholar] [CrossRef]
- Misbah, I.; Zhao, F.; Shih, W.-C. Symmetry Breaking-Induced Plasmonic Mode Splitting in Coupled Gold–Silver Alloy Nanodisk Array for Ultrasensitive RGB Colorimetric Biosensing. ACS Appl. Mater. Interfaces 2019, 11, 2273–2281. [Google Scholar] [CrossRef]
- Zhao, F.; Arnob, M.M.P.; Zenasni, O.; Li, J.; Shih, W.C. Far-field plasmonic coupling in 2-dimensional polycrystalline plasmonic arrays enables wide tunability with low-cost nanofabrication. Nanoscale Horiz. 2017, 2, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Kasani, S.; Zheng, P.; Wu, N. Tailoring Optical Properties of a Large-Area Plasmonic Gold Nanoring Array Pattern. J. Phys. Chem. C 2018, 122, 13443–13449. [Google Scholar] [CrossRef]
- Larsson, E.M.; Alegret, J.; Kǎll, M.; Sutherland, D.S. Sensing characteristics of NIR localized surface plasmon resonances in gold nanorings for application as ultrasensitive biosensors. Nano Lett. 2007, 7, 1256–1263. [Google Scholar] [CrossRef]
- Zhu, S.; Li, H.; Yang, M.; Pang, S.W. Label-free detection of live cancer cells and DNA hybridization using 3D multilayered plasmonic biosensor. Nanotechnology 2018, 29, 365503. [Google Scholar] [CrossRef]
- Li, C.; Li, Z.; Li, S.; Zhang, Y.; Sun, B.; Yu, Y.; Ren, H.; Jiang, S.; Yue, W. LSPR optical fiber biosensor based on a 3D composite structure of gold nanoparticles and multilayer graphene films. Opt. Express 2020, 28, 6071. [Google Scholar] [CrossRef] [PubMed]
- Auguié, B.; Barnes, W.L. Collective Resonances in Gold Nanoparticle Arrays. Phys. Rev. Lett. 2008, 101, 143902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kravets, V.G.; Schedin, F.; Kabashin, A.V.; Grigorenko, A.N. Sensitivity of collective plasmon modes of gold nanoresonators to local environment. Opt. Lett. 2010, 35, 956. [Google Scholar] [CrossRef] [Green Version]
- Kravets, V.G.; Schedin, F.; Grigorenko, A.N. Extremely narrow plasmon resonances based on diffraction coupling of localized plasmons in arrays of metallic nanoparticles. Phys. Rev. Lett. 2008, 101, 087403. [Google Scholar] [CrossRef] [PubMed]
- Danilov, A.; Tselikov, G.; Wu, F.; Kravets, V.G.; Ozerov, I.; Bedu, F.; Grigorenko, A.N.; Kabashin, A.V. Ultra-narrow surface lattice resonances in plasmonic metamaterial arrays for biosensing applications. Biosens. Bioelectron. 2018, 104, 102–112. [Google Scholar] [CrossRef] [Green Version]
- Zou, S.; Janel, N.; Schatz, G.C. Silver nanoparticle array structures that produce remarkably narrow plasmon lineshapes. J. Chem. Phys. 2004, 120, 10871–10875. [Google Scholar] [CrossRef]
- Markel, V.A. Comment on “Silver nanoparticle array structures that produce remarkably narrow plasmon line shapes” [J. Chem. Phys. 120, 10871 (2004)]. J. Chem. Phys. 2005, 122, 097101. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, S.R.K.; Schaafsma, M.C.; Berrier, A.; Gmez Rivas, J. Collective resonances in plasmonic crystals: Size matters. Phys. B: Condens. Matter 2012, 407, 4081–4085. [Google Scholar] [CrossRef] [Green Version]
- Ponomareva, E.; Volk, K.; Mulvaney, P.; Karg, M. Surface Lattice Resonances in Self-Assembled Gold Nanoparticle Arrays: Impact of Lattice Period, Structural Disorder, and Refractive Index on Resonance Quality. Langmuir 2020, 36, 13601–13612. [Google Scholar] [CrossRef] [PubMed]
- Li, X.X.; Zhang, T.; Yu, J.; Xing, C.; Li, X.X.; Cai, W.; Li, Y. Highly Selective and Sensitive Detection of Hydrogen Sulfide by the Diffraction Peak of Periodic Au Nanoparticle Array with Silver Coating. ACS Appl. Mater. Interfaces 2020, 12, 40702–40710. [Google Scholar] [CrossRef]
- Zou, S.; Schatz, G.C. Narrow plasmonic/photonic extinction and scattering line shapes for one and two dimensional silver nanoparticle arrays. J. Chem. Phys. 2004, 121, 12606–12612. [Google Scholar] [CrossRef]
- Zou, S.; Schatz, G.C. Theoretical studies of plasmon resonances in one-dimensional nanoparticle chains: Narrow lineshapes with tunable widths. Nanotechnology 2006, 17, 2813–2820. [Google Scholar] [CrossRef]
- Wang, B.; Yu, P.; Wang, W.; Zhang, X.; Kuo, H.C.; Xu, H.; Wang, Z.M. High-Q Plasmonic Resonances: Fundamentals and Applications. Adv. Opt. Mater. 2021, 9, 2001520. [Google Scholar] [CrossRef]
- Li, R.; Bourgeois, M.R.; Cherqui, C.; Guan, J.; Wang, D.; Hu, J.; Schaller, R.D.; Schatz, G.C.; Odom, T.W. Hierarchical Hybridization in Plasmonic Honeycomb Lattices. Nano Lett. 2019, 19, 6435–6441. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Shi, L.; Hong, M.; Li, H.; Li, D.; Liu, M. A surface plasmon resonance biosensor based on gold nanoparticle array. Opt. Commun. 2013, 298–299, 232–236. [Google Scholar] [CrossRef]
- Ahmed, T.; Paul, A.K.; Anower, M.S.; Razzak, S.M.A. Surface plasmon resonance biosensor based on hexagonal lattice dual-core photonic crystal fiber. Appl. Opt. 2019, 58, 8416. [Google Scholar] [CrossRef]
- Wu, T.; Shao, Y.; Wang, Y.; Cao, S.; Cao, W.; Zhang, F.; Liao, C.; He, J.; Huang, Y.; Hou, M.; et al. Surface plasmon resonance biosensor based on gold-coated side-polished hexagonal structure photonic crystal fiber. Opt. Express 2017, 25, 20313. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Butun, S.; Aydin, K. Ultranarrow band absorbers based on surface lattice resonances in nanostructured metal surfaces. ACS Nano 2014, 8, 8242–8248. [Google Scholar] [CrossRef]
- Chang, C.C.; Chen, C.P.; Wu, T.H.; Yang, C.H.; Lin, C.W.; Chen, C.Y. Gold nanoparticle-based colorimetric strategies for chemical and biological sensing applications. Nanomaterials 2019, 9, 861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iarossi, M.; Schiattarella, C.; Rea, I.; De Stefano, L.; Fittipaldi, R.; Vecchione, A.; Velotta, R.; Ventura, B. Della Colorimetric Immunosensor by Aggregation of Photochemically Functionalized Gold Nanoparticles. ACS Omega 2018, 3, 3805–3812. [Google Scholar] [CrossRef]
- Della Ventura, B.; Banchelli, M.; Funari, R.; Illiano, A.; De Angelis, M.; Taroni, P.; Amoresano, A.; Matteini, P.; Velotta, R. Biosensor surface functionalization by a simple photochemical immobilization of antibodies: Experimental characterization by mass spectrometry and surface enhanced Raman spectroscopy. Analyst 2019, 144, 6871–6880. [Google Scholar] [CrossRef]
- Funari, R.; Della Ventura, B.; Altucci, C.; Offenhäusser, A.; Mayer, D.; Velotta, R. Single Molecule Characterization of UV-Activated Antibodies on Gold by Atomic Force Microscopy. Langmuir 2016, 32, 8084–8091. [Google Scholar] [CrossRef]
- Oliverio, M.; Perotto, S.; Messina, G.C.; Lovato, L.; De Angelis, F. Chemical Functionalization of Plasmonic Surface Biosensors: A Tutorial Review on Issues, Strategies, and Costs. ACS Appl. Mater. Interfaces 2017, 9, 29394–29411. [Google Scholar] [CrossRef] [Green Version]
- Minopoli, A.; Sakač, N.; Lenyk, B.; Campanile, R.; Mayer, D.; Offenhäusser, A.; Velotta, R.; Della Ventura, B. LSPR-based colorimetric immunosensor for rapid and sensitive 17β-estradiol detection in tap water. Sens. Actuators B Chem. 2020, 308, 127699. [Google Scholar] [CrossRef]
- Habib, P.; Dreymueller, D.; Rösing, B.; Botung, H.; Slowik, A.; Zendedel, A.; Habib, S.; Hoffmann, S.; Beyer, C. Estrogen serum concentration affects blood immune cell composition and polarization in human females under controlled ovarian stimulation. J. Steroid Biochem. Mol. Biol. 2018, 178, 340–347. [Google Scholar] [CrossRef]
- Tian, J.-M.; Ran, B.; Zhang, C.-L.; Yan, D.-M.; LI, X.-H. Estrogen and progesterone promote breast cancer cell proliferation by inducing cyclin G1 expression. Braz. J. Med. Biol. Res. 2018, 51, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kidd, K.A.; Blanchfield, P.J.; Mills, K.H.; Palace, V.P.; Evans, R.E.; Lazorchak, J.M.; Flick, R.W. Collapse of a fish population after exposure to a synthetic estrogen. Proc. Natl. Acad. Sci. USA 2007, 104, 8897–8901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ventura, B.D.; Cennamo, M.; Minopoli, A.; Campanile, R.; Censi, S.B.; Terracciano, D.; Portella, G.; Velotta, R. Colorimetric Test for Fast Detection of SARS-CoV-2 in Nasal and Throat Swabs. ACS Sens. 2020, 5, 3043–3048. [Google Scholar] [CrossRef]
- Minopoli, A.; Scardapane, E.; Acunzo, A.; Campanile, R.; Della Ventura, B.; Velotta, R. Analysis of the optical response of a SARS-CoV-2-directed colorimetric immunosensor. AIP Adv. 2021, 11, 065319. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, L.; Wei, W.; Zhao, H.; Zhou, Z.; Zhang, Y.; Liu, S. Colorimetric detection of influenza A virus using antibody-functionalized gold nanoparticles. Analyst 2015, 140, 3989–3995. [Google Scholar] [CrossRef]
- Bosak, A.; Saraf, N.; Willenberg, A.; Kwan, M.W.C.; Alto, B.W.; Jackson, G.W.; Batchelor, R.H.; Nguyen-Huu, T.D.; Sankarapani, V.; Parks, G.D.; et al. Aptamer-gold nanoparticle conjugates for the colorimetric detection of arboviruses and vector mosquito species. RSC Adv. 2019, 9, 23752–23763. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Huo, D.; Jiang, H.; Dong, L.; Ma, Y.; Hou, C.; Fa, H.; Yang, M.; Luo, X.; Li, J.; et al. Highly Selective and Sensitive Colorimetric Sensor for Aminotriazole Residues in Vegetables and Fruits Using Glutathione Functionalized Gold Nanoparticles. J. Nanosci. Nanotechnol. 2017, 17, 4733–4739. [Google Scholar] [CrossRef]
- Yang, J.; Han, Y.; Zhang, R.; Zhang, R.; Li, J. Comparison of analytical sensitivity of SARS-CoV-2 molecular detection kits. Int. J. Infect. Dis. 2021, 111, 233–241. [Google Scholar] [CrossRef]
- Komiazyk, M.; Walory, J.; Kozinska, A.; Wasko, I.; Baraniak, A. Impact of the nucleic acid extraction method and the RT-qPCR assay on SARS-CoV-2 detection in low-viral samples. Diagnostics 2021, 11, 2247. [Google Scholar] [CrossRef] [PubMed]
- Pramanik, A.; Gao, Y.; Patibandla, S.; Mitra, D.; McCandless, M.G.; Fassero, L.A.; Gates, K.; Tandon, R.; Chandra Ray, P. The rapid diagnosis and effective inhibition of coronavirus using spike antibody attached gold nanoparticles. Nanoscale Adv. 2021, 3, 1588–1596. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Hosokawa, K.; Maeda, M. Rapid aggregation of gold nanoparticles induced by non-cross-linking DNA hybridization. J. Am. Chem. Soc. 2003, 125, 8102–8103. [Google Scholar] [CrossRef]
- Wang, G.; Akiyama, Y.; Takarada, T.; Maeda, M. Rapid Non-Crosslinking Aggregation of DNA-Functionalized Gold Nanorods and Nanotriangles for Colorimetric Single-Nucleotide Discrimination. Chem. A Eur. J. 2016, 22, 258–263. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Jiang, Q.; Wang, Y.; Yang, L.; Yu, P.; Mao, L. Real-time colorimetric assay of inorganic pyrophosphatase activity based on reversibly competitive coordination of Cu2+ between cysteine and pyrophosphate ion. Anal. Chem. 2013, 85, 9409–9415. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Xia, Y. Enzymatic Reaction Modulated Gold Nanorod End-to-End Self-Assembly for Ultrahigh Sensitively Colorimetric Sensing of Cholinesterase and Organophosphate Pesticides in Human Blood. Anal. Chem. 2015, 87, 8584–8591. [Google Scholar] [CrossRef]
- Kim, N.W.; Piatyszek, M.A.; Prowse, K.R.; Harley, C.B.; West, M.D.; Ho, P.L.C.; Coviello, G.M.; Wright, W.E.; Weinrich, S.L.; Shay, J.W. Specific association of human telomerase activity with immortal cells and cancer. Science 1994, 266, 2011–2015. [Google Scholar] [CrossRef]
- Shay, J.W.; Wright, W.E. Telomerase: A target for cancer therapeutics. Cancer Cell 2002, 2, 257–265. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wu, L.; Ren, J.; Qu, X. Visualizing human telomerase activity with primer-modified Au nanoparticles. Small 2012, 8, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wu, L.; Ren, J.; Qu, X. Visual detection of telomerase activity with a tunable dynamic range by using a gold nanoparticle probe-based hybridization protection strategy. Nanoscale 2014, 6, 1661–1666. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, S.; Pan, W.; Liang, Q.; Song, X. Exonuclease I manipulating primer-modified gold nanoparticles for colorimetric telomerase activity assay. Biosens. Bioelectron. 2016, 77, 144–148. [Google Scholar] [CrossRef] [PubMed]
- Aldewachi, H.S.; Woodroofe, N.; Turega, S.; Gardiner, P.H.E. Optimization of gold nanoparticle-based real-time colorimetric assay of dipeptidyl peptidase IV activity. Talanta 2017, 169, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.J.; Lee, D.I.; Kim, C.; Lee, K.; Lee, C.H.; Ahn, I.S. Gold nanoparticles-based colorimetric assay for cathepsin B activity and the efficiency of its inhibitors. Anal. Chem. 2014, 86, 3825–3833. [Google Scholar] [CrossRef] [PubMed]
- Moitra, P.; Alafeef, M.; Alafeef, M.; Alafeef, M.; Dighe, K.; Frieman, M.B.; Pan, D.; Pan, D.; Pan, D. Selective Naked-Eye Detection of SARS-CoV-2 Mediated by N Gene Targeted Antisense Oligonucleotide Capped Plasmonic Nanoparticles. ACS Nano 2020, 14, 7617–7627. [Google Scholar] [CrossRef]
- Cheraghi Shahi, S.; Dadmehr, M.; Korouzhdehi, B.; Tavassoli, A. A novel colorimetric biosensor for sensitive detection of aflatoxin mediated by bacterial enzymatic reaction in saffron samples. Nanotechnology 2021, 32, 505503. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Selegård, R.; Aili, D.; Liedberg, B. Peptide functionalized gold nanoparticles for colorimetric detection of matrilysin (MMP-7) activity. Nanoscale 2013, 5, 8973–8976. [Google Scholar] [CrossRef]
- Nossier, A.I.; Mohammed, O.S.; Fakhr El-deen, R.R.; Zaghloul, A.S.; Eissa, S. Gelatin-modified gold nanoparticles for direct detection of urinary total gelatinase activity: Diagnostic value in bladder cancer. Talanta 2016, 161, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Mizukami, S.; Kikuchi, K. Simple and real-time colorimetric assay for glycosidases activity using functionalized gold nanoparticles and its application for inhibitor screening. Anal. Chem. 2012, 84, 9089–9095. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Tang, Y.; Liu, J.; Jiang, L.; Huang, W.; Huo, F.W.; Tian, D. Colorimetric assay for heterogeneous-catalyzed lipase activity: Enzyme-regulated gold nanoparticle aggregation. J. Agric. Food Chem. 2015, 63, 39–42. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Kou, X.; Xu, Y.; Yang, D.; Miao, P. Colorimetric sensing strategy for heparin assay based on PDDA-induced aggregation of gold nanoparticles. Nanoscale Adv. 2019, 1, 486–489. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Bai, W.; Niu, S.; Zhu, C.; Yang, S.; Chen, A. Highly sensitive colorimetric detection of 17β-estradiol using split DNA aptamers immobilized on unmodified gold nanoparticles. Sci. Rep. 2014, 4, 7571. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.S.; Choo, K.H.; Lee, B.; Choi, S.J. The methods of identification, analysis, and removal of endocrine disrupting compounds (EDCs) in water. J. Hazard. Mater. 2009, 172, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Jung, H.S.; Matsuura, T.; Lee, H.Y.; Kawai, T.; Gu, M.B. Electrochemical detection of 17β-estradiol using DNA aptamer immobilized gold electrode chip. Biosens. Bioelectron. 2007, 22, 2525–2531. [Google Scholar] [CrossRef]
- Alsager, O.A.; Kumar, S.; Zhu, B.; Travas-Sejdic, J.; McNatty, K.P.; Hodgkiss, J.M. Ultrasensitive colorimetric detection of 17-estradiol: The effect of shortening dna aptamer sequences. Anal. Chem. 2015, 87, 4201–4209. [Google Scholar] [CrossRef] [PubMed]
- Qiao, L.; Wang, H.; He, J.; Yang, S.; Chen, A. Truncated affinity-improved aptamers for 17β-estradiol determination by AuNPs-based colorimetric aptasensor. Food Chem. 2021, 340, 128181. [Google Scholar] [CrossRef] [PubMed]
- Li, C.M.; Zhen, S.J.; Wang, J.; Li, Y.F.; Huang, C.Z. A gold nanoparticles-based colorimetric assay for alkaline phosphatase detection with tunable dynamic range. Biosens. Bioelectron. 2013, 43, 366–371. [Google Scholar] [CrossRef]
- Pan, Y.; Guo, M.; Nie, Z.; Huang, Y.; Peng, Y.; Liu, A.; Qing, M.; Yao, S. Colorimetric detection of apoptosis based on caspase-3 activity assay using unmodified gold nanoparticles. Chem. Commun. 2012, 48, 997–999. [Google Scholar] [CrossRef]
- He, Y.; Cheng, F.; Pang, D.-W.; Tang, H.-W. Colorimetric and visual determination of DNase I activity using gold nanoparticles as an indicator. Microchim. Acta 2017, 184, 101–106. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, J.; Jiang, J.; Yu, R. Enzyme-regulated unmodified gold nanoparticle aggregation: A label free colorimetric assay for rapid and sensitive detection of adenosine deaminase activity and inhibition. Chem. Commun. 2012, 48, 10996–10998. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; He, Y.; Xing, X.J.; Tan, D.D.; Lin, Y.; Pang, D.W.; Tang, H.W. A gold nanoparticle-based label free colorimetric aptasensor for adenosine deaminase detection and inhibition assay. Analyst 2015, 140, 1572–1577. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, H.; Chen, Z.; Wang, X.; Choo, J.; Chen, L. Plasmonic colorimetric sensors based on etching and growth of noble metal nanoparticles: Strategies and applications. Biosens. Bioelectron. 2018, 114, 52–65. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.M.; Jiao, L.; Cui, M.L.; Lin, L.P.; Wang, X.X.; Zheng, Z.Y.; Zhang, L.H.; Jiang, S.L. A highly sensitive non-aggregation colorimetric sensor for the determination of I- based on its catalytic effect on Fe3+ etching gold nanorods. Sens. Actuators B Chem. 2013, 188, 644–650. [Google Scholar] [CrossRef]
- Liu, J.M.; Wang, X.X.; Cui, M.L.; Lin, L.P.; Jiang, S.L.; Jiao, L.; Zhang, L.H. A promising non-aggregation colorimetric sensor of AuNRs-Ag+ for determination of dopamine. Sens. Actuators B Chem. 2013, 176, 97–102. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, Z.; Qu, C.; Chen, L. Highly sensitive visual detection of copper ions based on the shape-dependent LSPR spectroscopy of gold nanorods. Langmuir 2014, 30, 3625–3630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, H.; Wang, E. Nanomaterials with enzyme-like characteristics (nanozymes): Next-generation artificial enzymes. Chem. Soc. Rev. 2013, 42, 6060–6093. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Ni, D.; Rosenkrans, Z.T.; Huang, P.; Yan, X.; Cai, W. Nanozyme: New horizons for responsive biomedical applications. Chem. Soc. Rev. 2019, 48, 3683–3704. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huang, D.; Lai, C.; Qin, L.; Zeng, G.; Xu, P.; Li, B.; Yi, H.; Zhang, M. Peroxidase-Like Activity of Smart Nanomaterials and Their Advanced Application in Colorimetric Glucose Biosensors. Small 2019, 15, 1900133. [Google Scholar] [CrossRef]
- Wu, J.; Wang, X.; Wang, Q.; Lou, Z.; Li, S.; Zhu, Y.; Qin, L.; Wei, H. Nanomaterials with enzyme-like characteristics (nanozymes): Next-generation artificial enzymes (II). Chem. Soc. Rev. 2019, 48, 1004–1076. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Zhuang, J.; Nie, L.; Zhang, J.; Zhang, Y.; Gu, N.; Wang, T.; Feng, J.; Yang, D.; Perrett, S.; et al. Intrinsic peroxidase-like activity of ferromagnetic nanoparticles. Nat. Nanotechnol. 2007, 2, 577–583. [Google Scholar] [CrossRef]
- Lu, C.; Liu, X.; Li, Y.; Yu, F.; Tang, L.; Hu, Y.; Ying, Y. Multifunctional Janus Hematite-Silica Nanoparticles: Mimicking Peroxidase-Like Activity and Sensitive Colorimetric Detection of Glucose. ACS Appl. Mater. Interfaces 2015, 7, 15395–15402. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Yan, Y.; Xiang, D.; Zhang, C.; Xian, Y. Magnetic Fe3S4 nanoparticles with peroxidase-like activity, and their use in a photometric enzymatic glucose assay. Microchim. Acta 2016, 183, 625–631. [Google Scholar] [CrossRef]
- Zhang, H.; Han, L.; Li, F. A universal one-pot assay strategy based on bio-inorganic cascade catalysts for different analytes by changing pH-dependent activity of enzymes on enzyme mimics. Sens. Actuators B Chem. 2019, 286, 460–467. [Google Scholar] [CrossRef]
- Han, L.; Liu, P.; Zhang, H.; Li, F.; Liu, A. Phage capsid protein-directed MnO 2 nanosheets with peroxidase-like activity for spectrometric biosensing and evaluation of antioxidant behaviour. Chem. Commun. 2017, 53, 5216–5219. [Google Scholar] [CrossRef]
- Liang, M.; Yan, X. Nanozymes: From New Concepts, Mechanisms, and Standards to Applications. Acc. Chem. Res. 2019, 52, 2190–2200. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, M.; Cao, W.; Wang, X.; Wang, Q.; Li, S.; Wei, H. Light-Responsive Metal-Organic Framework as an Oxidase Mimic for Cellular Glutathione Detection. Anal. Chem. 2019, 91, 8170–8175. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, J.; Su, D.; Zhang, Y.; Lu, H.; Yan, X.; Bai, J.; Gao, Y.; Lu, G. The DNA controllable peroxidase mimetic activity of MoS2nanosheets for constructing a robust colorimetric biosensor. Nanoscale 2020, 12, 19420–19428. [Google Scholar] [CrossRef] [PubMed]
- Rashtbari, S.; Dehghan, G.; Amini, M. An ultrasensitive label-free colorimetric biosensor for the detection of glucose based on glucose oxidase-like activity of nanolayered manganese-calcium oxide. Anal. Chim. Acta 2020, 1110, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Ma, Z.; Li, P.; Lu, Q.; Liu, M.; Li, H.; Zhang, Y.; Yao, S. Bifunctional colorimetric biosensors via regulation of the dual nanoenzyme activity of carbonized FeCo-ZIF. Sens. Actuators B Chem. 2019, 290, 357–363. [Google Scholar] [CrossRef]
- Ren, H.; Liu, X.; Yan, L.; Cai, Y.; Liu, C.; Zeng, L.; Liu, A. Ocean green tide derived hierarchical porous carbon with bi-enzyme mimic activities and their application for sensitive colorimetric and fluorescent biosensing. Sens. Actuators B Chem. 2020, 312, 127979. [Google Scholar] [CrossRef]
- Wu, C.W.; Unnikrishnan, B.; Tseng, Y.T.; Wei, S.C.; Chang, H.T.; Huang, C.C. Mesoporous manganese oxide/manganese ferrite nanopopcorns with dual enzyme mimic activities: A cascade reaction for selective detection of ketoses. J. Colloid Interface Sci. 2019, 541, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Savanur, M.A.; Srivastava, S.; D’Silva, P.; Mugesh, G. A Redox Modulatory Mn 3 O 4 Nanozyme with Multi-Enzyme Activity Provides Efficient Cytoprotection to Human Cells in a Parkinson’s Disease Model. Angew. Chemie Int. Ed. 2017, 56, 14267–14271. [Google Scholar] [CrossRef] [PubMed]
- He, S.-B.; Chen, R.-T.; Wu, Y.-Y.; Wu, G.-W.; Peng, H.-P.; Liu, A.-L.; Deng, H.-H.; Xia, X.-H.; Chen, W. Improved enzymatic assay for hydrogen peroxide and glucose by exploiting the enzyme-mimicking properties of BSA-coated platinum nanoparticles. Microchim. Acta 2019, 186, 778. [Google Scholar] [CrossRef]
- Bhagat, S.; Srikanth Vallabani, N.V.; Shutthanandan, V.; Bowden, M.; Karakoti, A.S.; Singh, S. Gold core/ceria shell-based redox active nanozyme mimicking the biological multienzyme complex phenomenon. J. Colloid Interface Sci. 2018, 513, 831–842. [Google Scholar] [CrossRef]
- Zhao, L.; Wu, Z.; Liu, G.; Lu, H.; Gao, Y.; Liu, F.; Wang, C.; Cui, J.; Lu, G. High-Activity Mo, S co-doped carbon quantum dot nanozyme-based cascade colorimetric biosensor for sensitive detection of cholesterol. J. Mater. Chem. B 2019, 7, 7042–7051. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yan, L.; Ren, H.; Cai, Y.; Liu, C.; Zeng, L.; Gao, J.; Liu, A. Facile synthesis of magnetic hierarchical flower-like Co3O4 spheres: Mechanism, excellent tetra-enzyme mimics and their colorimetric biosensing applications. Biosens. Bioelectron. 2020, 165, 112342. [Google Scholar] [CrossRef]
- Ferhan, A.R.; Guo, L.; Zhou, X.; Chen, P.; Hong, S.; Kim, D.H. Solid-phase colorimetric sensor based on gold nanoparticle-loaded polymer brushes: Lead detection as a case study. Anal. Chem. 2013, 85, 4094–4099. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Chang, H.T.; Shiang, Y.C.; Hung, Y.L.; Chiang, C.K.; Huang, C.C. Colorimetric assay for lead ions based on the leaching of gold nanoparticles. Anal. Chem. 2009, 81, 9433–9439. [Google Scholar] [CrossRef]
- Lee, Y.F.; Huang, C.C. Colorimetric assay of lead ions in biological samples using a nanogold-based membrane. ACS Appl. Mater. Interfaces 2011, 3, 2747–2754. [Google Scholar] [CrossRef]
- Xia, Y.; Ye, J.; Tan, K.; Wang, J.; Yang, G. Colorimetric visualization of glucose at the submicromole level in serum by a homogenous silver nanoprism-glucose oxidase system. Anal. Chem. 2013, 85, 6241–6247. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, Z.; Cheng, F.; Zhang, Y.; Chen, L. Highly sensitive on-site detection of glucose in human urine with naked eye based on enzymatic-like reaction mediated etching of gold nanorods. Biosens. Bioelectron. 2017, 89, 932–936. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, Z.; Chen, L. Ultrasensitive Visual Sensing of Molybdate Based on Enzymatic-like Etching of Gold Nanorods. Langmuir 2015, 31, 9253–9259. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jiao, J.; Wei, Y.; Wang, D.; Yang, C.; Xu, Z. Plasmonic Colorimetric Biosensor for Sensitive Exosome Detection via Enzyme-Induced Etching of Gold Nanobipyramid@MnO 2 Nanosheet Nanostructures. Anal. Chem. 2020, 92, 15244–15252. [Google Scholar] [CrossRef] [PubMed]
- Li, X.B.; Zhang, Z.R.; Schluesener, H.J.; Xu, S.Q. Role of exosomes in immune regulation. J. Cell. Mol. Med. 2006, 10, 364–375. [Google Scholar] [CrossRef]
- Park, J.E.; Tan, H.S.; Datta, A.; Lai, R.C.; Zhang, H.; Meng, W.; Lim, S.K.; Sze, S.K. Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol. Cell. Proteom. 2010, 9, 1085–1099. [Google Scholar] [CrossRef] [Green Version]
- Hough, K.P.; Chanda, D.; Duncan, S.R.; Thannickal, V.J.; Deshane, J.S. Exosomes in immunoregulation of chronic lung diseases. Allergy Eur. J. Allergy Clin. Immunol. 2017, 72, 534–544. [Google Scholar] [CrossRef]
- Tao, Z.; Gao, P.; Hoffman, D.W.; Liu, H.W. Domain C of human poly(ADP-ribose) polymerase-1 is important for enzyme activity and contains a novel zinc-ribbon motif. Biochemistry 2008, 47, 5804–5813. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xu, E.; Xu, C.; Wei, W. Colorimetric method for PARP-1 detection based on preventing AuNRs from etching by molybdate. Sens. Actuators B Chem. 2020, 325, 128806. [Google Scholar] [CrossRef]
- Liang, J.; Yao, C.; Li, X.; Wu, Z.; Huang, C.; Fu, Q.; Lan, C.; Cao, D.; Tang, Y. Silver nanoprism etching-based plasmonic ELISA for the high sensitive detection of prostate-specific antigen. Biosens. Bioelectron. 2015, 69, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.C.; Chen, T.H.; Tseng, W.L.; Lin, C.H. Novel core etching technique of gold nanoparticles for colorimetric dopamine detection. Analyst 2012, 137, 5352–5357. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Lin, Y.; Guo, L.; Qiu, B.; Chen, G.; Yang, H.H.; Lin, Z. A universal multicolor immunosensor for semiquantitative visual detection of biomarkers with the naked eyes. Biosens. Bioelectron. 2017, 87, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, P.; Mao, X.; Fu, W.; Liu, C. Seed-mediated growth of bimetallic nanoparticles as an effective strategy for sensitive detection of Vitamin C. Sens. Actuators B Chem. 2016, 231, 95–101. [Google Scholar] [CrossRef]
- Lin, T.; Li, Z.; Song, Z.; Chen, H.; Guo, L.; Fu, F.; Wu, Z. Visual and colorimetric detection of p-aminophenol in environmental water and human urine samples based on anisotropic growth of Ag nanoshells on Au nanorods. Talanta 2016, 148, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Shaban, S.M.; Moon, B.S.; Kim, D.H. Selective and sensitive colorimetric detection of p-aminophenol in human urine and paracetamol drugs based on seed-mediated growth of silver nanoparticles. Environ. Technol. Innov. 2021, 22, 101517. [Google Scholar] [CrossRef]
- Wang, Y.; Zeng, Y.; Fu, W.; Zhang, P.; Li, L.; Ye, C.; Yu, L.; Zhu, X.; Zhao, S. Seed-mediated growth of Au@Ag core-shell nanorods for the detection of ellagic acid in whitening cosmetics. Anal. Chim. Acta 2018, 1002, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Jackson, A.A.; Rotello, V.M.; Nugen, S.R. Colorimetric Detection of Escherichia coli Based on the Enzyme-Induced Metallization of Gold Nanorods. Small 2016, 12, 2469–2475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.; Yang, J.; Wang, H.F.; Wang, Z.; Huang, X.; Wang, Z.; Niu, G.; Hight Walker, A.R.; Chen, X. Glucose oxidase-catalyzed growth of gold nanoparticles enables quantitative detection of attomolar cancer biomarkers. Anal. Chem. 2014, 86, 5800–5806. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Ouyang, W.; Xie, P.; Lin, Y.; Qiu, B.; Lin, Z.; Chen, G.; Guo, L. Highly Uniform Gold Nanobipyramids for Ultrasensitive Colorimetric Detection of Influenza Virus. Anal. Chem. 2017, 89, 1617–1623. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, J.; Ekman, J.M.; Kenis, P.J.A.; Lu, Y. DNA-mediated control of metal nanoparticle shape: One-pot synthesis and cellular uptake of highly stable and functional gold nanoflowers. Nano Lett. 2010, 10, 1886–1891. [Google Scholar] [CrossRef]
- Soh, J.H.; Lin, Y.; Rana, S.; Ying, J.Y.; Stevens, M.M. Colorimetric Detection of Small Molecules in Complex Matrixes via Target-Mediated Growth of Aptamer-Functionalized Gold Nanoparticles. Anal. Chem. 2015, 87, 7644–7652. [Google Scholar] [CrossRef] [PubMed]


















| Transducer | Matrix | Analytes | LR | S | LOD | Ref. |
|---|---|---|---|---|---|---|
| AgNPs | whole human blood | dengue NS1 | 0.5–50 μg/mL | 9 nm/(μg/mL) | 0.06 μg/mL | [48] |
| ELISA (Eagle Bioscience) | serum, plasma, cell culture supernatants | dengue NS1 | 1.56–100 ng/mL | 1.56 ng/mL | ||
| Au hemispheres | human serum | CitH3 | 10−4–10 ng/mL | 300 nm/RIU | 0.87 pg/mL | [49] |
| ELISA (Cayman Chemical) | serum, plasma, cell culture supernatants | CitH3 | 0.15–10 ng/mL | 0.1 ng/mL | ||
| Au hemispheres | buffer serum | anti-human IgG | n/a | 211 nm/RIU | 1 μM | [50] |
| L-shape Au stripes | buffer solution | miRNA-let-7a | 1 fM–100 pM | n/a | 13 fM | [54] |
| ELISA (BioVendor) | whole blood, cell culture lysates | hsa-miR-let-7a-5p | 1.25–40 pM | 0.42 pM | ||
| Au nanomushrooms | buffer solution | β-giardin gene | 10 nM–1μM | 83–108 nm/RIU | 5 nM | [55] |
| Al bowtie | n/a | n/a | n/a | 500 nm/RIU | n/a | [57] |
| Au nanoislands | aqueous solution | DNA | n/a | 120 nm/RIU | 10 nM | [58] |
| Au nanodisks | buffer solution | ssDNA | 10–170 nM * | 210 nm/RIU | 10 nM | [59] |
| Au nanodisks | water | ARC DNA | 10–1010 fM ** | 227 nm/RIU | 10 fM | [32] |
| Transducer | Matrix | Analytes | LR | S | LOD | Ref. |
|---|---|---|---|---|---|---|
| Core–shell Au nanocones | buffer solution | DNA | 1–105 pM | 417.5 nm/RIU | 161 fM | [33] |
| Au crossed-bowties | n/a | n/a | n/a | 1753 nm/RIU | n/a | [69] |
| Au–Ag alloy nanodisks | buffer solution | streptavidin-biotin | 0.1–10 nM | 344 nm/RIU | 0.1 nM | [77] |
| Au film on nanopillars | ultrapure water | DNA | 0.1–104 pM | 382–442 nm/RIU | 70 fM | [81] |
| AuNP multilayer | buffer solution | T-DNA | 0.1–100 nM | 1251.44 nm/RIU | 0.1 nM | [82] |
| Transducer | Matrix | Analytes | LR | S | LOD | Ref. |
|---|---|---|---|---|---|---|
| rectangular lattice of AuNPs | PBS | streptavidin–biotin | n/a | 190–200 * | n/a | [86] |
| 400–420 ** | ||||||
| hexagonal lattice of AuNPs | human blood plasma | H2S | 0–12 μM *** | n/a | 0.79 μM | [91] |
| Rectangular lattice of Au cubes placed on quartz pillars | refractive index sensing | n/a | 596.7 nm/RIU | n/a | [96] | |
| hexagonal lattice dual-core photonic crystal fiber | refractive index sensing | 1.33–1.40 | 16,000 nm/RIU | n/a | [97] | |
| Au-coated side-polished hexagonal structure photonic crystal fiber | refractive index sensing | 1.36–1.44 | 21,700 nm/RIU | n/a | [98] | |
| Au nanoring/nanowire array | refractive index sensing | n/a | 900 nm/RIU | n/a | [99] | |
| Transducer | Analyte | Matrix | LR | LOD | Ref. |
|---|---|---|---|---|---|
| anti-human IgG f-AuNPs | human IgG | ultrapure water | 50–500 ng/mL | <100 ng/mL | [101] |
| anti-E2 f-AuNPs | E2 | tap water | 4–10 pg/mL | 3 pg/mL | [105] |
| ELISA (Cayman Chemical) | E2 | cit plasma, EDTA plasma, hep plasma, serum, urine | 0.61–104 pg/mL | 6 pg/mL | |
| glutathione f-AuNPs | aminotriazole | buffer solution | 0.59–21.40 μM | 0.27 μM | [113] |
| SARS-CoV-2 anti-envelope-, anti-membrane-, and anti-spike-f-AuNPs | SARS-CoV-2 | Universal Transport Medium | 30–7 Ct | 36.5 Ct | [109] |
| RT-PCR | SARS-CoV-2 | nasopharyngeal swab | 16–106 copies/mL | 16 copies/mL | [114,115] |
| anti-hemagglutinin f-AuNPs | IAV | PBS | 40–320 HAU/mL | 31.2 HAU/mL | [111] |
| ELISA (Sino Biological) | influenza H1N1 hemagglutinin | supernatant | 31.25–2000 pg/mL | 31.25 pg/mL | |
| CFA0335 aptamer-conjugated AuNPs | salivary mosquito proteins | buffer solution | 10–250 ng * | 10 ng | [112] |
| CFA0335 aptamer-conjugated AuNPs | ZIKV-E | buffer solution | n/a | 0.4 nM | [112] |
| ELISA (Sino Biological) | ZIKV-E | serum, supernatant | 187.5–12,000 pg/mL | 99.72 pg/mL | |
| anti-spike-f-AuNPs | COVID-19 spike recombinant antigen | buffer solution | 1–10 ng/mL ** | 1 ng/mL | [116] |
| pseudo-SARS-CoV-2 | buffer solution | 1000–5000 virus particles/mL ** | 1000 virus particles/mL |
| Transducer | Analyte | Matrix | LR | LOD | IC50 (*) | Ref. |
|---|---|---|---|---|---|---|
| cysteine-stabilized AuNPs | PPase | water | 0.025–0.4 U | 0.010 U | n/a | [119] |
| NaF | water | 1–50 μM ** | 1 μM ** | 7.1 μM (0.2 U) | ||
| AuNRs | cholinesterase | buffer solution | 0.042–8.4 μU/mL | 0.018 μU/mL | n/a | [120] |
| ELISA (Abbexa) | cholinesterase | serum, plasma | 0.625–40 ng/mL | 0.38 ng/mL | ||
| AuNRs | organophosphate pesticides | buffer solution | 0.12–40 pM | 0.039 pM | 1.2 pM (42 μU/mL) *** | [120] |
| TS-AuNPs | telomerase extracts from HeLa cells | buffer solution | 1–8 cells/μL | 1 cell/ μL | n/a | [123] |
| ELISA (MyBioSource) | telomerase | serum, plasma, tissue homogenates | 0.625–20 ng/mL | 0.625 ng/mL | ||
| TC-AuNPs | telomerase extracts from HeLa cells | buffer solution | 100–4000 cells (2 nM [AuNPs]) | 100 cells (2 nM AuNPs) | n/a | [124] |
| 500–10,000 cells (5 nM [AuNPs]) | 500 cells ** (5 nM AuNPs) | n/a | ||||
| ELISA (MyBioSource) | telomerase | serum, plasma, tissue homogenates | 0.625–20 ng/mL | 0.625 ng/mL | ||
| TC-AuNPs | Curcumin | buffer solution | 1–20 μM ** | 1 μM ** | 2.8 μM (4000 cells) | [124] |
| TP-AuNPs | telomerase extracted from HL-60 cancer cells | buffer solution | 0–200 cells/mL | 29 cells/mL | n/a | [125] |
| 100 cells/mL (naked-eye) | ||||||
| ELISA (MyBioSource) | telomerase | serum, plasma, tissue homogenates | 0.625–20 ng/mL | 0.625 ng/mL | ||
| TP-AuNPs | BIBR1532 | buffer solution | 20–500 nM ** | 20 nM ** | 50 nM (300 cells/mL) | [125] |
| Transducer | Analyte | Matrix | LR | LOD | IC50 (*) | Ref. |
|---|---|---|---|---|---|---|
| ASO-capped AuNPs | N-gene | nasopharyngeal swab | 0.2–3 ng/μL | 0.18 ng/μL | n/a | [128] |
| RT-PCR | SARS-CoV-2 | nasopharyngeal swab | 16–106 copies/mL | 16 copies/mL | [114,115] | |
| GPDC-modified AuNPs | DPP-IV | 50 mM Tris buffer (pH 8.3) | 0–30 U/L | 1.2 U/L | n/a | [126] |
| ELISA (Thermo Fisher Scientific) | DPP-IV | plasma, serum, supernatant | 0.41–100 ng/mL | 0.45 ng/mL | ||
| VPED-DC-modified AuNPs | DPP-IV | 50 mM Tris buffer (pH 8.3) | 0–12 U/L | 1.5 U/L | n/a | [126] |
| ELISA (Thermo Fisher Scientific) | DPP-IV | plasma, serum, supernatant | 0.41–100 ng/mL | 0.45 ng/mL | ||
| AuNPs@gelatin | AFB1 | bacterial supernatant containing gelatinase enzyme | 10–140 pg/mL | 4 pg/mL | n/a | [129] |
| ELISA (Elabscience) | AFB1 | cereals, corn skin, wheat bran, edible oil, peanut, biscuits, beer, wine, soy sauce, vinegar | 0.03–0.48 ng/mL | 0.03 ng/mL | ||
| DBDY-Gly-Phe-MTPA-AuNPs | cathepsin B | phosphate buffer | 10–50 nM ** | 10 nM (1 h reaction) | n/a | [127] |
| phosphate buffer | 5–50 nM ** | 5 nM (2 h reaction) | n/a | |||
| ELISA (Abcam) | cathepsin B | cell culture supernatant, cell lysate, plasma, serum, tissue extracts | 156–10,000 pg/mL | 156 pg/mL | ||
| DBDY-Gly-Phe-MTPA-AuNPs | leupeptin | phosphate buffer | 0–0.5 μM ** | n/a | 0.11 μM (50 nM) | [127] |
| DBDY-Gly-Phe-MTPA-AuNPs | antipain | phosphate buffer | 0–2 μM ** | n/a | 0.48 μM (50 nM) | [127] |
| DBDY-Gly-Phe-MTPA-AuNPs | chymostatin | phosphate buffer | 0–5 μM ** | n/a | 1.78 μM (50 nM) | [127] |
| JR2EC-functionalized AuNPs | MMP-7 | PBS (pH 7.4) | 0–2 μg/mL | 0.1 μg/mL | n/a | [130] |
| ELISA (Thermo Fisher Scientific) | MMP-7 | supernatant | 0.15–100 ng/mL | 0.15 ng/mL | ||
| JR2EC-functionalized AuNPs | MMP inhibitor II | PBS (pH 7.4) | n/a | n/a | 30 nM (2 μg/mL) | [130] |
| Gelatin-modified AuNPs | MMP2/MMP9 | 10 mM 6-MCH | 1.85–148 ng/mL | 1.85 ng/mL | n/a | [131] |
| ELISA (Thermo Fisher Scientific) | MMP2 | plasma, serum, supernatant | 0.78–50 ng/mL | <1 ng/mL | ||
| ELISA (Thermo Fisher Scientific) | MMP9 | plasma, serum, supernatant | 0.23–15.0 ng/mL | 0.05 ng/mL | ||
| Gal-Lip-AuNPs | β-galactosidase | 50 mM PBS (pH 7.4) | 0.02–2.5 μM ** | 9.2 nM (20 min reaction time) | n/a | [132] |
| 2.9 nM (90 min reaction time) | ||||||
| ELISA (MyBioSource) | human β-galactosidase | serum, plasma, tissue homogenates | 0.312–20 mIU/mL | 0.312 mIU/mL | ||
| Gal-Lip-AuNPs | D-galactal | 50 mM PBS (pH 7.4) | 50–1000 μM ** | 50 μM ** | 482 μM (1.0 μM) | [132] |
| Glc-Lip-AuNPs | β-glucosidase | 50 mM PBS (pH 7.4) | 0.02–2.5 μM ** | 22.3 nM (20 min reaction time) | n/a | [132] |
| 9.8 nM (90 min reaction time) | ||||||
| ELISA (Bioassay Technology Laboratory) | human β-glucosidase | serum, plasma, cell culture supernatants | 20–6000 ng/L | 20 ng/L | ||
| Glc-Lip-AuNPs | castanospermine | 50 mM PBS (pH 7.4) | 10–400 μM ** | 10 μM ** | 46.4 μM (1.0 μM) | [132] |
| Tween 20-AuNPs | lipase | 0.5 M PBS, 2 M NaCl (pH 6.5) | 0.15–1.80 mg/mL | 0.028 mg/mL | n/a | [133] |
| ELISA (Antibodies-online) | lipase | gallus plasma, serum, tissue homogenates | 0.15–10 ng/mL | 0.15 ng/mL |
| Transducer | Analyte | Matrix | LR | LOD | Ref. |
|---|---|---|---|---|---|
| aptamer-capped AuNPs | E2 | ultrapure water | 0.1–105 ng/mL | 0.1 ng/mL | [135] |
| ELISA (Cayman Chemical) | E2 | cit plasma, EDTA plasma, hep plasma, serum, urine | 0.61–104 pg/mL | 6 pg/mL | |
| aptamer-capped AuNPs | E2 | filter river water | 200–800 pM | 200 pM | [138] |
| E2 | spiked rat urine | 50–800 nM | 5 nM | ||
| ELISA (Cayman Chemical) | E2 | cit plasma, EDTA plasma, hep plasma, serum, urine | 0.61–104 pg/mL | 6 pg/mL | |
| aptamer-capped AuNPs | E2 | ultrapure water | 0.02–10 μg/mL | 0.02 μg/mL * | [139] |
| ELISA (Cayman Chemical) | E2 | cit plasma, EDTA plasma, hep plasma, serum, urine | 0.61–104 pg/mL | 6 pg/mL | |
| AuNP@CTAB/ATP | ALP | Tris-HCl buffer | 100–600 unit/mL | 10 unit/mL | [140] |
| AuNP@CTAB/ATP/Ca2+ | ALP | Tris-HCl buffer | 5.0–100 unit/mL | 3.5 unit/mL | |
| AuNP@CTAB/ATP/Pb2+ | ALP | Tris-HCl buffer | 0.2–20 unit/mL | 0.1 unit/mL | |
| ELISA (MyBioSource) | ALP | human serum, plasma, tissue homogenates | 0.156–10 ng/mL | 0.156 ng/mL | |
| AuNPs@Asp-Glu-Val-Asp,DEVD | caspase-3 | GR-8 solution | 0.01–0.15 μg/mL | 0.005 μg/mL | [141] |
| ELISA (Thermo Fisher Scientific) | caspase-3 | human serum, plasma, supernatant | 0.16–10.0 ng/mL | 0.16 ng/mL | |
| AuNPs@dsDNA | DNase 1 | Tris-HCl buffer | 0–35 unit/L | 7.1 unit/L | [142] |
| citrate-capped AuNPs | adenosine deaminase | buffer solution | 0–15 U/L | 0.8227 U/L | [143] |
| ELISA (MyBioSource) | human adenosine deaminase | serum, urine, tissue homogenates | 1.56–100 mIU/mL | 1.56 mIU/mL | |
| citrate-capped AuNPs | adenosine deaminase | HEPES buffer | 0–21 U/L | 1.526 U/L | [144] |
| ELISA (MyBioSource) | human adenosine deaminase | serum, urine, tissue homogenates | 1.56–100 mIU/mL | 1.56 mIU/mL |
| Transducer | Enzyme-Mimic | Analyte | Matrix (pH) | LR | LOD | Ref. |
|---|---|---|---|---|---|---|
| FeS2NPs | peroxidase-like | H2O2 | acetate buffer (4.0) | 2–80 μM | 0.91 μM | [34] |
| GSH | acetate buffer (4.0) | 0.2–35 μM | 0.15 μM | |||
| ELISA (MyBioSource) | GSH | serum, plasma, cell culture supernatant | 1.56–100 μg/mL | 1.56 μg/mL | ||
| DNA/MoS2-NSs | peroxidase-like | carcinoembryonic antigen | acetate buffer (4.0) | 50–1000 ng/mL | 50 ng/mL | [160] |
| ELISA (Abcam) | carcinoembryonic antigen | serum, plasma, cell culture supernatant | 0.343–250 ng/mL | 0.343 ng/mL | ||
| NL-MnCaO2 | glucose oxidase-like | glucose | acetate buffer (5.0) | 18.3–421 μM | 23.86 μM | [161] |
| human serum | 0–82.3 μM | 6.12 μM | ||||
| ELISA (Abcam) | glucose | cell culture supernatant, urine, serum, plasma | 1–10,000 µM | 1 µM | ||
| FeCo@C | oxidase-like | hydroquinone | acetate buffer (3.6) | 1–30 μM | 0.8 μM | [162] |
| peroxidase-like | H2O2 | acetate buffer (4.4) | 1–240 μM | 1 μM | ||
| Mo-CQDs | peroxidase-like | H2O2 | acetate buffer (4.0) | 5–50 μM | - | [168] |
| cholesterol | acetate buffer (4.0) | 10–600 μM | 7 μM | |||
| ELISA (LifeSpan BioSciences) | human cholesterol | plasma, serum | 0.5–10 mM | 0.5 mM | ||
| magnetic hierarchical Co3O4 nanoflowers | oxidase-like | ACP | acetate buffer (4.5) | 0.1–25 U/L | 0.062 U/L | [169] |
| peroxidase-like | H2O2 | air | 4–400 μM | 2 μM | ||
| ELISA (Abcam) | ACP | cell culture media, cell lysate, plasma, serum, tissue extracts, urine | 0.001–1 mM | 0.001 mM |
| Transducer | Analyte | Matrix | LR | LOD | Ref. |
|---|---|---|---|---|---|
| AuNRs | molybdate | acetate buffer solution | 3–70 nM | 1 nM | [175] |
| AuNBPs | telomerase extracts from HeLa cells | buffer solution | 5–1000 cells | 1 cell 20 cells (naked-eye) | [35] |
| ELISA (MyBioSource) | telomerase | serum, plasma, tissue homogenates | 0.625–20 ng/mL | 0.625 ng/mL | |
| AuNBP@MnO2NSs | exosome | buffer solution | 6.8 × 104–1.5 × 105 particles/μL (low-AR) | 2.20 × 103 particles/μL (low-AR) | [176] |
| 8.5 × 102–8.5 × 104 particles/μL (high-AR) | 1.35 × 102 particles/μL (high-AR) | ||||
| AuNRs | PARP-1 | buffer solution | 0.05–2 U | 0.02 U | [181] |
| ELISA (MyBioSource) | PARP-1 | serum, plasma, tissue homogenates, cell lysates | 15.6–1000 pg/mL | 15.6 pg/mL | |
| Ab-GOx-MBs/AgNPRs | PSA | foetal bovine serum | 10–105 fg/mL | 4.1 fg/mL | [182] |
| ELISA (Abcam) | human PSA | cell culture supernatant, cit plasma, EDTA plasma, hep plasma, serum, urine | 39–2500 pg/mL | 39 pg/mL | |
| DMAP-AuNPs | dopamine | deionized water | 10–100 nM | 5 nM | [183] |
| ELISA (Abcam) | dopamine | cell culture supernatant, plasma, serum, tissue homogenate | 1.56–100 ng/mL | 1.56 ng/mL | |
| AuNRs | carcinoembryonic antigen | human serum | 2.5–60 ng/mL * | 2.5 ng/mL | [184] |
| ELISA (Abcam) | carcinoembryonic antigen | serum, plasma, cell culture supernatant | 0.343–250 ng/mL | 0.343 ng/mL | |
| AuNRs | prostate specific antigen | human serum | 75–1275 pg/mL * | 75 pg/mL | [184] |
| ELISA (Abcam) | human PSA | cell culture supernatant, cit plasma, EDTA plasma, hep plasma, serum, urine | 39–2500 pg/mL | 39 pg/mL |
| Transducer | Analyte | Matrix | LR | LOD | Ref. |
|---|---|---|---|---|---|
| AgNPs | pAP | human urine | 0–85 μM | 0.32 μM | [187] |
| ELISA (Abcam) | human pAP | cell culture media, cell culture supernatant, cit plasma, EDTA plasma, hep plasma, serum, urine | 37.5–2400 pg/mL | 37.5 pg/mL | |
| AuNRs | EA | buffer solution | 0–20 μM | 0.040 μM | [188] |
| AuNRs | β-gal | buffer solution | 0–2 nM | 0.128 nM | [189] |
| ELISA (LifeSpan BioSciences) | β-gal | plasma, serum | 0.313–20 mU/mL | 0.313 mU/mL | |
| AuNRs | E. coli | buffer solution | 104–106 CFU/mL * | 1 × 104 CFU/mL | [189] |
| ELISA (Immuno-Biological Laboratories) | E. coli O157:H7 | food, water | 6.25 × 103–4 × 105 CFU/mL | 6.25 × 103 CFU/mL | |
| AuNBs | H5N1 | buffer solution | 1–2.5 × 103 pg/mL | 1 pg/mL | [191] |
| AuNPs | ochratoxin | buffer solution | 12.5–125 nM * | 1 nM | [193] |
| ELISA (MyBioSource) | ochratoxin A | cereal, feed | 0.1–8.1 ppb | 0.1 ppb | |
| AuNPs | cocaine | synthetic urine sample | 0.1–100 nM * | 1 nM | [193] |
| ELISA (MyBioSource) | human cocaine-regulated transcript protein | serum, plasma, tissue homogenates | 15.6–1000 pg/mL | 15.6 pg/mL | |
| AuNPs | E2 | synthetic saliva sample | 0.01–104 nM * | 0.2 nM | [193] |
| ELISA (Cayman Chemical) | E2 | cit plasma, EDTA plasma, hep plasma, serum, urine | 0.61–104 pg/mL | 6 pg/mL |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Acunzo, A.; Scardapane, E.; De Luca, M.; Marra, D.; Velotta, R.; Minopoli, A. Plasmonic Nanomaterials for Colorimetric Biosensing: A Review. Chemosensors 2022, 10, 136. https://doi.org/10.3390/chemosensors10040136
Acunzo A, Scardapane E, De Luca M, Marra D, Velotta R, Minopoli A. Plasmonic Nanomaterials for Colorimetric Biosensing: A Review. Chemosensors. 2022; 10(4):136. https://doi.org/10.3390/chemosensors10040136
Chicago/Turabian StyleAcunzo, Adriano, Emanuela Scardapane, Maria De Luca, Daniele Marra, Raffaele Velotta, and Antonio Minopoli. 2022. "Plasmonic Nanomaterials for Colorimetric Biosensing: A Review" Chemosensors 10, no. 4: 136. https://doi.org/10.3390/chemosensors10040136
APA StyleAcunzo, A., Scardapane, E., De Luca, M., Marra, D., Velotta, R., & Minopoli, A. (2022). Plasmonic Nanomaterials for Colorimetric Biosensing: A Review. Chemosensors, 10(4), 136. https://doi.org/10.3390/chemosensors10040136