Programmable, Universal DNAzyme Amplifier Supporting Pancreatic Cancer-Related miRNAs Detection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Apparatus
2.2. Materials
2.3. Polypropylene Gel Electrophoresis Analysis
2.4. Reaction Kinetics of the Substrate-Cleaving Performance of the DNAzyme Amplifier
2.5. Investigation of Influencing Factors on the Performance of DNAzyme Amplifier
2.6. Fluorescence Assay of miR-10b
2.7. Selective Detection of miR-10b
2.8. Detection of miR-10b in Human Serum Samples
3. Results and Discussion
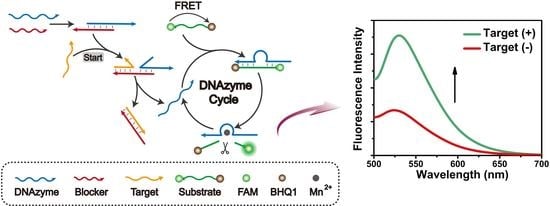
3.1. Mechanism of the DNAzyme Amplifier
3.2. Feasibility of the DNAzyme Amplifier
3.3. Reaction Kinetics of the Substrate-Cleaving Performance of the DNAzyme Amplifier
3.4. Effects on the Performance of DNAzyme Amplifier
3.5. Detection Capability of MiR-10b by the DNAzyme Amplifier
3.6. Practical Detectability of the DNAzyme Amplifier
3.7. Universal Detection of the DNAzyme Amplifier for miR-21
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jun, I.; Hirochika, T.; Ippei, M.; Sadaki, A.; Tadahiro, G.; Sachio, T.; Yoshihide, N.; Azusa, Y.; Takuya, M.; Yuki, U.; et al. Second primary pancreatic ductal carcinoma in the remnant pancreas after pancreatectomy for pancreatic ductal carcinoma: High cumulative incidence rates at 5 years after pancreatectomy. Pancreatology 2016, 16, 615–620. [Google Scholar]
- Shinji, I.; Yasutsugu, S.; Tsunehiro, M.; Toshiji, T.; Takayuki, N.; Hiroyuki, T.; Satoru, T.; Masamichi, K.; Taichi, T.; Ayana, I.; et al. Two case reports of resectable cancer in the remnant pancreas after pancreatectomy for invasive ductal carcinoma of the pancreas. J. Clim. 2018, 103, 542–547. [Google Scholar]
- Lu, T.X.; Rothenberg, M.E. MicroRNA. J. Allergy Clin. Immunol. 2018, 141, 1202–1207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fathi, M.; Ghafouri-Fard, S.; Abak, A.; Taheri, M. Emerging roles of miRNAs in the development of pancreatic cancer. Biomed. Pharmacother. 2021, 141, 111914. [Google Scholar] [CrossRef]
- Nakata, K.; Ohuchida, K.; Mizumoto, K.; Kayashima, T.; Ikenaga, N.; Sakai, H.; Lin, C.; Fujita, H.; Otsuka, T.; Aishima, S.; et al. MicroRNA-10b is overexpressed in pancreatic cancer, promotes its invasiveness, and correlates with a poor prognosis. Surgery 2011, 150, 916–922. [Google Scholar] [CrossRef]
- Schwarzkopf, M.; Pierce, N.A. Multiplexed miRNA northern blots via hybridization chain reaction. Nucleic Acids Res. 2016, 44, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucchinetti, E.; Zaugg, M. RNA sequencing. Anesthesiology 2020, 133, 976–978. [Google Scholar] [CrossRef]
- Babapoor, S.; Horwich, M.; Wu, R.; Levinson, S.; Gandhi, M.; Makkar, H.; Kristjansson, A.; Chang, M.; Dadras, S.S. MicroRNA in situ hybridization for miR-211 detection as an ancillary test in melanoma diagnosis. Mod. Pathol. 2016, 29, 461–475. [Google Scholar] [CrossRef] [Green Version]
- Pritchard, C.C.; Cheng, H.H.; Tewari, M. MicroRNA profiling: Approaches and considerations. Nat. Rev. Genet. 2012, 13, 358–369. [Google Scholar] [CrossRef]
- Androvic, P.; Valihrach, L.; Elling, J.; Sjoback, R.; Kubista, M. Two-tailed RT-qPCR: A novel method for highly accurate miRNA quantification. Nucleic Acids Res. 2017, 45, e144. [Google Scholar] [CrossRef]
- Balkourani, G.; Brouzgou, A.; Archonti, M.; Papandrianos, N.; Song, S.; Tsiakaras, P. Emerging materials for the electrochemical detection of COVID-19. J. Electroanal. Chem. 2021, 893, 115289. [Google Scholar] [CrossRef] [PubMed]
- Negahdary, M.; Angnes, L. Application of electrochemical biosensors for the detection of microRNAs (miRNAs) related to cancer. Coord. Chem. Rev. 2022, 464, 214565. [Google Scholar] [CrossRef]
- Su, J.; Wang, D.; Lena, N.; Shen, J.; Zhao, Z.; Dou, Y.; Peng, T.; Shi, J.; Sanjay, M.; Fan, C.; et al. Multicolor gold–silver nano-mushrooms as ready-to-use SERS probes for Ultrasensitive and multiplex DNA/miRNA detection. Anal. Chem. 2017, 89, 2531–2538. [Google Scholar] [CrossRef]
- Ki, J.; Lee, H.Y.; Son, H.Y.; Huh, Y.M.; Haam, S. Sensitive plasmonic detection of miR-10b in biological samples using enzyme-assisted target recycling and developed LSPR probe. ACS Appl. Mater. Interfaces 2019, 11, 18923–18929. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Ma, C.; Zhu, J.-J. DNA technology-assisted signal amplification strategies in electrochemiluminescence bioanalysis. J. Anal. Test. 2021, 5, 95–111. [Google Scholar] [CrossRef]
- Jiang, Y.J.; Yang, X.J.; Wang, J.; Li, Y.F.; Li, C.M.; Huang, C.Z. Soft nanoball-encapsulated carbon dots for reactive oxygen species scavenging and the highly sensitive chemiluminescent assay of nucleic acid biomarkers. Analyst 2021, 146, 7187–7193. [Google Scholar] [CrossRef]
- Ki, J.; Jang, E.; Han, S.; Shin, M.K.; Kang, B.; Huh, Y.M.; Haam, S. Instantaneous pH-boosted functionalization of stellate gold nanoparticles for intracellular imaging of miRNA. ACS Appl. Mater. Interfaces 2017, 9, 17702–17709. [Google Scholar] [CrossRef]
- Li, X.; Chen, S.; Liu, Q.; Luo, Y.; Sun, X. Hexagonal boron nitride nanosheet as an effective nanoquencher for the fluorescence detection of microRNA. Chem. Commun. 2021, 57, 8039–8042. [Google Scholar] [CrossRef]
- Cheng, Y.Y.; Xie, Y.F.; Li, C.M.; Li, Y.F.; Huang, C.Z. Förster resonance energy transfer-based soft nanoballs for specific and amplified detection of microRNAs. Anal. Chem. 2019, 91, 11023–11029. [Google Scholar] [CrossRef]
- Moros, M.; Kyriazi, M.-E.; El-Sagheer, A.H.; Brown, T.; Tortiglione, C.; Kanaras, A.G. DNA-coated gold nanoparticles for the detection of mRNA in live hydra vulgaris animals. ACS Appl. Mater. Interfaces 2019, 11, 13905–13911. [Google Scholar] [CrossRef]
- Chen, M.; Duan, R.; Xu, S.; Duan, Z.; Yuan, Q.; Xia, F.; Huang, F. Photoactivated DNA walker based on DNA nanoflares for signal-amplified microRNA Imaging in single living cells. Anal. Chem. 2021, 93, 16264–16272. [Google Scholar] [CrossRef] [PubMed]
- Xing, C.; Chen, S.; Lin, Q.; Lin, Y.; Wang, M.; Wang, J.; Lu, C. An aptamer-tethered DNA origami amplifier for sensitive and accurate imaging of intracellular microRNA. Nanoscale 2022, 14, 1327–1332. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Guo, Z. Special topic: Novel sensing materials and their applications in analytical chemistry. J. Anal. Test. 2021, 5, 1–2. [Google Scholar] [CrossRef]
- Robertson, N.M.; Hizir, M.S.; Balcioglu, M.; Wang, R.; Yavuz, M.S.; Yumak, H.; Ozturk, B.; Sheng, J.; Yigit, M.V. Discriminating a single nucleotide difference for enhanced miRNA detection using tunable graphene and oligonucleotide nanodevices. Langmuir 2015, 31, 9943–9952. [Google Scholar] [CrossRef]
- Yoo, B.; Kavishwar, A.; Ghosh, S.K.; Barteneva, N.; Yigit, M.V.; Moore, A.; Medarova, Z. Detection of miRNA expression in intact cells using activatable sensor oligonucleotides. Chem. Biol. 2014, 21, 199–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, K.X.; Qiao, Z.J.; Song, W.L.; Bi, S. DNA nanotechnology for multimodal synergistic theranostics. J. Anal. Test. 2021, 5, 112–129. [Google Scholar] [CrossRef]
- Quan, K.; Li, J.; Wang, J.; Xie, N.; Wei, Q.; Tang, J.; Yang, X.; Wang, K.; Huang, J. Dual-microRNA-controlled double-amplified cascaded logic DNA circuits for accurate discrimination of cell subtypes. Chem. Sci. 2019, 10, 1442–1449. [Google Scholar] [CrossRef] [Green Version]
- Deng, R.; Tang, L.; Tian, Q.; Wang, Y.; Lin, L.; Li, J. Toehold-initiated rolling circle amplification for visualizing individual microRNAs in situ in single cells. Angew. Chem. 2014, 126, 2421–2425. [Google Scholar] [CrossRef]
- Abdullah Al-Maskri, A.A.; Ye, J.; Talap, J.; Hu, H.; Sun, L.; Yu, L.; Cai, S.; Zeng, S. Reverse transcription-based loop-mediated isothermal amplification strategy for real-time miRNA detection with phosphorothioated probes. Anal. Chim. Acta 2020, 1126, 1–6. [Google Scholar] [CrossRef]
- Jin, F.; Xu, D. A fluorescent microarray platform based on catalytic hairpin assembly for MicroRNAs detection. Anal. Chim. Acta 2021, 1173, 338666. [Google Scholar] [CrossRef]
- Duan, R.; Zuo, X.; Wang, S.; Quan, X.; Chen, D.; Chen, Z.; Jiang, L.; Fan, C.; Xia, F. Lab in a tube: Ultrasensitive detection of microRNAs at the single-cell level and in breast cancer patients using quadratic isothermal amplification. J. Am. Chem. Soc. 2013, 135, 4604–4607. [Google Scholar] [CrossRef] [PubMed]
- Aihua, T.; Yu, L.; Jian, G. Sensitive SERS detection of lead ions via DNAzyme based quadratic signal amplification. Talanta 2017, 171, 185–189. [Google Scholar]
- Liu, S.; Wei, W.; Sun, X.; Wang, L. Ultrasensitive electrochemical DNAzyme sensor for lead ion based on cleavage-induced template-independent polymerization and alkaline phosphatase amplification. Biosens. Bioelectron. 2016, 83, 33–38. [Google Scholar] [CrossRef]
- Liu, H.; Yang, Q.; Peng, R.; Kuai, H.; Lyu, Y.; Pan, X.; Liu, Q.; Tan, W. Artificial signal feedback network mimicking cellular adaptivity. J. Am. Chem. Soc. 2019, 141, 6458–6461. [Google Scholar] [CrossRef]
- Wu, Z.; Fan, H.; Satyavolu, N.S.R.; Wang, W.; Lake, R.; Jiang, J.-H.; Lu, Y. Imaging endogenous metal ions in living cells using a DNAzyme—Catalytic hairpin assembly probe. Angew. Chem. Int. Ed. 2017, 56, 8721–8725. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Luan, Y.; Xiong, M.; Zhang, J.; Lake, R.; Lu, Y. DNAzyme amplified aptasensing platform for ochratoxin a detection using a personal glucose meter. ACS Appl. Mater. Interfaces 2021, 13, 9472–9481. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Luo, M.; Wang, L.; Li, C.; Hu, S.; Yu, X.; Yuan, P.; Wu, Z.S. Stimuli-responsive autonomous-motion molecular machine for sensitive simultaneous fluorescence imaging of intracellular microRNAs. Anal. Chem. 2021, 93, 9869–9877. [Google Scholar] [CrossRef] [PubMed]






| Sample | Add/nM | Found/nM Mean a ± SD b | Recovery/% (n = 3) | RSD/% (n = 3) |
|---|---|---|---|---|
| 1 | 20 | 19 ± 0.43 | 94.12 | 2.3 |
| 2 | 40 | 43 ± 1.39 | 106.73 | 3.3 |
| 3 | 60 | 62 ± 0.86 | 103.63 | 1.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nie, K.; Jiang, Y.; Wang, N.; Wang, Y.; Li, D.; Zhan, L.; Huang, C.; Li, C. Programmable, Universal DNAzyme Amplifier Supporting Pancreatic Cancer-Related miRNAs Detection. Chemosensors 2022, 10, 276. https://doi.org/10.3390/chemosensors10070276
Nie K, Jiang Y, Wang N, Wang Y, Li D, Zhan L, Huang C, Li C. Programmable, Universal DNAzyme Amplifier Supporting Pancreatic Cancer-Related miRNAs Detection. Chemosensors. 2022; 10(7):276. https://doi.org/10.3390/chemosensors10070276
Chicago/Turabian StyleNie, Kunhan, Yongjian Jiang, Na Wang, Yajun Wang, Di Li, Lei Zhan, Chengzhi Huang, and Chunmei Li. 2022. "Programmable, Universal DNAzyme Amplifier Supporting Pancreatic Cancer-Related miRNAs Detection" Chemosensors 10, no. 7: 276. https://doi.org/10.3390/chemosensors10070276
APA StyleNie, K., Jiang, Y., Wang, N., Wang, Y., Li, D., Zhan, L., Huang, C., & Li, C. (2022). Programmable, Universal DNAzyme Amplifier Supporting Pancreatic Cancer-Related miRNAs Detection. Chemosensors, 10(7), 276. https://doi.org/10.3390/chemosensors10070276