Development of Anti-OSCAR Antibodies for the Treatment of Osteoarthritis
Abstract
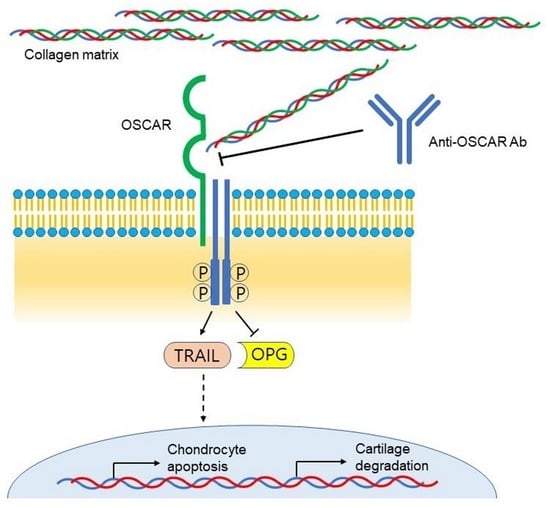
:1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Production of OSCAR-Fc Fusion Proteins
2.3. Identification of OSCAR-Binding Antibodies
2.4. Production of Anti-OSCAR mAbs
2.5. Chondrocyte-Based Assay to Identify Antibodies That Inhibit OSCAR In Vitro
2.6. RNA Isolation and Quantitative RT-PCR
2.7. Primary Articular Chondrocyte Assay to Identify Antibodies That Inhibit OSCAR In Vitro
2.8. Osteoclast Differentiation Assay to Identify Antibodies That Inhibit OSCAR In Vitro
2.9. Affinity Maturation of the B4 Clone by Light Chain Shuffling
2.10. Affinity Maturation of the D11 Clone by CDR-H3 Randomization
2.11. Measurement of Antibody Affinity by Biolayer Interferometry (BLI)
2.12. Experimental OA in Mice
2.13. Histology, Immunohistochemistry, and TUNEL Fluorescence Microscopy of Joint Sections
2.14. Quantitation and Statistical Analysis
3. Results
3.1. Generation of Anti-OSCAR Antibodies by Phage Display
3.2. In Vitro Functional Characterization of Anti-OSCAR Antibodies
3.3. Affinity Maturation of Anti-OSCAR Antibodies
3.4. Affinities of the Optimized Antibodies for Human and Mouse OSCAR-Fc
3.5. In Vivo Efficacy of the Affinity-Matured Anti-OSCAR Antibodies
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lories, R.J.; Luyten, F.P. The bone-cartilage unit in osteoarthritis. Nat. Rev. Rheumatol. 2011, 7, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Loeser, R.F.; Goldring, S.R.; Scanzello, C.R.; Goldring, M.B. Osteoarthritis: A disease of the joint as an organ. Arthritis Rheum. 2012, 64, 1697–1707. [Google Scholar] [CrossRef] [PubMed]
- Akkiraju, H.; Nohe, A. Role of Chondrocytes in Cartilage Formation, Progression of Osteoarthritis and Cartilage Regeneration. J. Dev. Biol. 2015, 3, 177–192. [Google Scholar] [CrossRef]
- Hwang, H.S.; Kim, H.A. Chondrocyte Apoptosis in the Pathogenesis of Osteoarthritis. Int. J. Mol. Sci. 2015, 16, 26035–26054. [Google Scholar] [CrossRef]
- Troeberg, L.; Nagase, H. Proteases involved in cartilage matrix degradation in osteoarthritis. Biochim. Biophys. Acta 2012, 1824, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Kim, J.; Ryu, J.H.; Oh, H.; Chun, C.H.; Kim, B.J.; Min, B.H.; Chun, J.S. Hypoxia-inducible factor-2alpha is a catabolic regulator of osteoarthritic cartilage destruction. Nat. Med. 2010, 16, 687–693. [Google Scholar] [CrossRef]
- Pettenuzzo, S.; Arduino, A.; Belluzzi, E.; Pozzuoli, A.; Fontanella, C.G.; Ruggieri, P.; Salomoni, V.; Majorana, C.; Berardo, A. Biomechanics of Chondrocytes and Chondrons in Healthy Conditions and Osteoarthritis: A Review of the Mechanical Characterisations at the Microscale. Biomedicines 2023, 11, 1942. [Google Scholar] [CrossRef]
- Kapoor, M.; Martel-Pelletier, J.; Lajeunesse, D.; Pelletier, J.P.; Fahmi, H. Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat. Rev. Rheumatol. 2011, 7, 33–42. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, X.; Wang, S.; Jing, Y.; Su, J. Subchondral bone microenvironment in osteoarthritis and pain. Bone Res. 2021, 9, 20. [Google Scholar] [CrossRef]
- Zhu, X.; Chan, Y.T.; Yung, P.S.H.; Tuan, R.S.; Jiang, Y. Subchondral Bone Remodeling: A Therapeutic Target for Osteoarthritis. Front. Cell Dev. Biol. 2020, 8, 607764. [Google Scholar] [CrossRef]
- Belluzzi, E.; Todros, S.; Pozzuoli, A.; Ruggieri, P.; Carniel, E.L.; Berardo, A. Human Cartilage Biomechanics: Experimental and Theoretical Approaches towards the Identification of Mechanical Properties in Healthy and Osteoarthritic Conditions. Processes 2023, 11, 1014. [Google Scholar] [CrossRef]
- Karsdal, M.A.; Michaelis, M.; Ladel, C.; Siebuhr, A.S.; Bihlet, A.R.; Andersen, J.R.; Guehring, H.; Christiansen, C.; Bay-Jensen, A.C.; Kraus, V.B. Disease-modifying treatments for osteoarthritis (DMOADs) of the knee and hip: Lessons learned from failures and opportunities for the future. Osteoarthr. Cartil. 2016, 24, 2013–2021. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Jeong, S.; Kim, H.; Kang, D.; Lee, J.; Kang, S.B.; Kim, J.H. Disease-modifying therapeutic strategies in osteoarthritis: Current status and future directions. Exp. Mol. Med. 2021, 53, 1689–1696. [Google Scholar] [CrossRef]
- Park, D.R.; Kim, J.; Kim, G.M.; Lee, H.; Kim, M.; Hwang, D.; Lee, H.; Kim, H.S.; Kim, W.; Park, M.C.; et al. Osteoclast-associated receptor blockade prevents articular cartilage destruction via chondrocyte apoptosis regulation. Nat. Commun. 2020, 11, 4343. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Takami, M.; Rho, J.; Josien, R.; Choi, Y. A novel member of the leukocyte receptor complex regulates osteoclast differentiation. J. Exp. Med. 2002, 195, 201–209. [Google Scholar] [CrossRef]
- Nemeth, K.; Schoppet, M.; Al-Fakhri, N.; Helas, S.; Jessberger, R.; Hofbauer, L.C.; Goettsch, C. The role of osteoclast-associated receptor in osteoimmunology. J. Immunol. 2011, 186, 13–18. [Google Scholar] [CrossRef]
- Barrow, A.D.; Raynal, N.; Andersen, T.L.; Slatter, D.A.; Bihan, D.; Pugh, N.; Cella, M.; Kim, T.; Rho, J.; Negishi-Koga, T.; et al. OSCAR is a collagen receptor that costimulates osteoclastogenesis in DAP12-deficient humans and mice. J. Clin. Investig. 2011, 121, 3505–3516. [Google Scholar] [CrossRef]
- Haywood, J.; Qi, J.; Chen, C.C.; Lu, G.; Liu, Y.; Yan, J.; Shi, Y.; Gao, G.F. Structural basis of collagen recognition by human osteoclast-associated receptor and design of osteoclastogenesis inhibitors. Proc. Natl. Acad. Sci. USA 2016, 113, 1038–1043. [Google Scholar] [CrossRef]
- Zhou, L.; Hinerman, J.M.; Blaszczyk, M.; Miller, J.L.; Conrady, D.G.; Barrow, A.D.; Chirgadze, D.Y.; Bihan, D.; Farndale, R.W.; Herr, A.B. Structural basis for collagen recognition by the immune receptor OSCAR. Blood 2016, 127, 529–537. [Google Scholar] [CrossRef]
- Koga, T.; Inui, M.; Inoue, K.; Kim, S.; Suematsu, A.; Kobayashi, E.; Iwata, T.; Ohnishi, H.; Matozaki, T.; Kodama, T.; et al. Costimulatory signals mediated by the ITAM motif cooperate with RANKL for bone homeostasis. Nature 2004, 428, 758–763. [Google Scholar] [CrossRef]
- Merck, E.; Gaillard, C.; Gorman, D.M.; Montero-Julian, F.; Durand, I.; Zurawski, S.M.; Menetrier-Caux, C.; Carra, G.; Lebecque, S.; Trinchieri, G.; et al. OSCAR is an FcRgamma-associated receptor that is expressed by myeloid cells and is involved in antigen presentation and activation of human dendritic cells. Blood 2004, 104, 1386–1395. [Google Scholar] [CrossRef] [PubMed]
- Nedeva, I.R.; Vitale, M.; Elson, A.; Hoyland, J.A.; Bella, J. Role of OSCAR Signaling in Osteoclastogenesis and Bone Disease. Front. Cell Dev. Biol. 2021, 9, 641162. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.M.; Park, H.; Lee, S.Y. Roles of osteoclast-associated receptor in rheumatoid arthritis and osteoarthritis. Jt. Bone Spine 2022, 89, 105400. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.Y.; Kang, K.J.; Chung, J.E.; Shim, H. Construction of a large synthetic human scFv library with six diversified CDRs and high functional diversity. Mol. Cells 2009, 27, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Kim, J.; Kang, S.; Kim, W.; Shim, H. A Novel Human scFv Library with Non-Combinatorial Synthetic CDR Diversity. PLoS ONE 2015, 10, e0141045. [Google Scholar] [CrossRef]
- Gosset, M.; Berenbaum, F.; Thirion, S.; Jacques, C. Primary culture and phenotyping of murine chondrocytes. Nat. Protoc. 2008, 3, 1253–1260. [Google Scholar] [CrossRef]
- Choi, H.K.; Kim, T.H.; Jhon, G.J.; Lee, S.Y. Reactive oxygen species regulate M-CSF-induced monocyte/macrophage proliferation through SHP1 oxidation. Cell Signal 2011, 23, 1633–1639. [Google Scholar] [CrossRef]
- Itzstein, C.; van ‘t Hof, R.J. Osteoclast formation in mouse co-cultures. Methods Mol. Biol. 2012, 816, 177–186. [Google Scholar] [CrossRef]
- Bai, X.; Jang, M.; Lee, N.J.; Nguyen, T.T.H.; Jung, M.; Hwang, J.Y.; Shim, H. A Novel Synthetic Antibody Library with Complementarity-Determining Region Diversities Designed for an Improved Amplification Profile. Int. J. Mol. Sci. 2022, 23, 6255. [Google Scholar] [CrossRef]
- Glasson, S.S.; Blanchet, T.J.; Morris, E.A. The surgical destabilization of the medial meniscus (DMM) model of osteoarthritis in the 129/SvEv mouse. Osteoarthr. Cartil. 2007, 15, 1061–1069. [Google Scholar] [CrossRef]
- Culley, K.L.; Dragomir, C.L.; Chang, J.; Wondimu, E.B.; Coico, J.; Plumb, D.A.; Otero, M.; Goldring, M.B. Mouse models of osteoarthritis: Surgical model of posttraumatic osteoarthritis induced by destabilization of the medial meniscus. Methods Mol. Biol. 2015, 1226, 143–173. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Huang, L.; Welch, I.; Norley, C.; Holdsworth, D.W.; Beier, F.; Cai, D. Early Changes of Articular Cartilage and Subchondral Bone in The DMM Mouse Model of Osteoarthritis. Sci. Rep. 2018, 8, 2855. [Google Scholar] [CrossRef] [PubMed]
- Glasson, S.S.; Chambers, M.G.; Van Den Berg, W.B.; Little, C.B. The OARSI histopathology initiative—Recommendations for histological assessments of osteoarthritis in the mouse. Osteoarthr. Cartil. 2010, 18 (Suppl. S3), S17–S23. [Google Scholar] [CrossRef] [PubMed]
- Newton, P.T.; Staines, K.A.; Spevak, L.; Boskey, A.L.; Teixeira, C.C.; Macrae, V.E.; Canfield, A.E.; Farquharson, C. Chondrogenic ATDC5 cells: An optimised model for rapid and physiological matrix mineralisation. Int. J. Mol. Med. 2012, 30, 1187–1193. [Google Scholar] [CrossRef]
- Temu, T.M.; Wu, K.Y.; Gruppuso, P.A.; Phornphutkul, C. The mechanism of ascorbic acid-induced differentiation of ATDC5 chondrogenic cells. Am. J. Physiol. Endocrinol. Metab. 2010, 299, E325–E334. [Google Scholar] [CrossRef] [PubMed]
- Ko, B.K.; Choi, S.; Cui, L.G.; Lee, Y.H.; Hwang, I.S.; Kim, K.T.; Shim, H.; Lee, J.S. Affinity Maturation of Monoclonal Antibody 1E11 by Targeted Randomization in CDR3 Regions Optimizes Therapeutic Antibody Targeting of HER2-Positive Gastric Cancer. PLoS ONE 2015, 10, e0134600. [Google Scholar] [CrossRef]
- Gram, H.; Marconi, L.A.; Barbas, C.F., 3rd; Collet, T.A.; Lerner, R.A.; Kang, A.S. In vitro selection and affinity maturation of antibodies from a naive combinatorial immunoglobulin library. Proc. Natl. Acad. Sci. USA 1992, 89, 3576–3580. [Google Scholar] [CrossRef]
- Weitzner, B.D.; Dunbrack, R.L., Jr.; Gray, J.J. The origin of CDR H3 structural diversity. Structure 2015, 23, 302–311. [Google Scholar] [CrossRef]
- Crotti, T.N.; Dharmapatni, A.A.; Alias, E.; Zannettino, A.C.; Smith, M.D.; Haynes, D.R. The immunoreceptor tyrosine-based activation motif (ITAM)-related factors are increased in synovial tissue and vasculature of rheumatoid arthritic joints. Arthritis Res. Ther. 2012, 14, R245. [Google Scholar] [CrossRef]




| Gene | Strand | Primer Sequence | Origin |
|---|---|---|---|
| Oscar | Sense | 5′-AGGGAAACCTCATCCGTTT-3′ | Mouse |
| Antisense | 5′-TGCTGTGCCAATCACAAGTA-3′ | ||
| Mmp3 | Sense | 5′-TCCTGATGTTGGTGGCTTCAG-3′ | Mouse |
| Antisense | 5′-TGTTTATTGTTGCTGCCCAT-3′ | ||
| Mmp13 | Sense | 5′-CCTTGAACGTCATCATCAGG-3′ | Mouse |
| Antisense | 5′-TGTTTATTGTTGCTGCCCAT-3′ | ||
| Epas1 | Sense | 5′-CGAGAAGAACGACGTGGTGTTC-3′ | Mouse |
| Antisense | 5′-GTGAAGGCGGGCAGGCTCC-3′ | ||
| Tnfsf10 | Sense | 5′-CCTCTCGGAAAGGGCATTC-3′ | Mouse |
| Antisense | 5′-TCCTGCTCGATGACCAGCT-3′ | ||
| Tnfrsf11b | Sense | 5′-CAGAGCGAAACACAGTTTG-3′ | Mouse |
| Antisense | 5′-CACACAGGGTGACATCTATTC-3′ | ||
| β-Actin | Sense | 5′-TGGAATCCTGTGGCATCCATGAAAC-3′ | Mouse |
| Antisense | 5′-TAAAACGCAGCTCAGTAACAGTCCG-3′ |
| Clone | CDR-H3 Sequence |
|---|---|
| D11 (parent) | AKAGFTGGHFDI |
| D11-3-B2 | AKAGNWGGHFDI |
| D11-3-F2 | AKAGNFGGHFDI |
| D11-3-F9 | AKAGLFGGHFDI |
| D11-4-B9 | AKAGIPG-HFDI |
| D11-5-F8 | AKAG—FTHFDI |
| D11-6-C10 | AKAG—FSMFDI |
| EC50 (nM) | D11 | D11-B9 | B4 | B4K-C11 | B4L-E2 |
|---|---|---|---|---|---|
| hOSCAR | >1000 | 36.8 | 590 | 6.74 | 1.33 |
| mOSCAR | 151 | 0.93 | 414 | 1.30 | 7.20 |
| Ligand | Parameters | D11 | D11-B9 | B4 | B4K-C11 | B4L-E2 |
|---|---|---|---|---|---|---|
| hOSCAR | kon (M−1s−1) | 9.13 × 104 | 1.01 × 105 | 8.99 × 104 | 1.54 × 105 | 1.38 × 105 |
| koff (s−1) | 5.12 × 10−4 | 4.15 × 10−5 | 2.92 × 10−4 | 3.37 × 10−5 | 1.21 × 10−5 | |
| KD (nM) | 5.61 | 0.41 | 3.25 | 0.22 | 0.088 | |
| mOSCAR | kon (M−1s−1) | 9.13 × 104 | 9.13 × 105 | 9.13 × 104 | 9.13 × 105 | 9.13 × 105 |
| koff (s−1) | 2.54 × 10−4 | 2.59 × 10−5 | 3.89 × 10−4 | 6.79 × 10−5 | 2.47 × 10−5 | |
| KD (nM) | 2.87 | 0.22 | 4.61 | 0.40 | 0.23 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, G.M.; Park, D.R.; Nguyen, T.T.H.; Kim, J.; Kim, J.; Sohn, M.-H.; Lee, W.-K.; Lee, S.Y.; Shim, H. Development of Anti-OSCAR Antibodies for the Treatment of Osteoarthritis. Biomedicines 2023, 11, 2844. https://doi.org/10.3390/biomedicines11102844
Kim GM, Park DR, Nguyen TTH, Kim J, Kim J, Sohn M-H, Lee W-K, Lee SY, Shim H. Development of Anti-OSCAR Antibodies for the Treatment of Osteoarthritis. Biomedicines. 2023; 11(10):2844. https://doi.org/10.3390/biomedicines11102844
Chicago/Turabian StyleKim, Gyeong Min, Doo Ri Park, Thi Thu Ha Nguyen, Jiseon Kim, Jihee Kim, Myung-Ho Sohn, Won-Kyu Lee, Soo Young Lee, and Hyunbo Shim. 2023. "Development of Anti-OSCAR Antibodies for the Treatment of Osteoarthritis" Biomedicines 11, no. 10: 2844. https://doi.org/10.3390/biomedicines11102844
APA StyleKim, G. M., Park, D. R., Nguyen, T. T. H., Kim, J., Kim, J., Sohn, M. -H., Lee, W. -K., Lee, S. Y., & Shim, H. (2023). Development of Anti-OSCAR Antibodies for the Treatment of Osteoarthritis. Biomedicines, 11(10), 2844. https://doi.org/10.3390/biomedicines11102844