A Biosafety Study of Human Umbilical Cord Blood Mononuclear Cells Transduced with Adenoviral Vector Carrying Human Vascular Endothelial Growth Factor cDNA In Vitro
Abstract
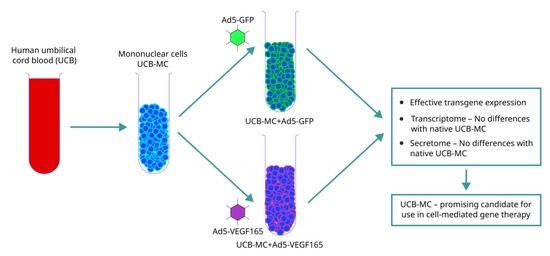
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Preparation of Umbilical Cord Blood Mononuclear Cells
2.3. Adenoviral Transduction of UCB-MCs
2.4. Flow Cytometry and Fluorescence Microscopy
2.5. Quantitative Reverse Transcription PCR
2.6. Western Blotting
2.7. Enzyme-Linked Immunosorbent Assay
2.8. Multiplex Secretome Profiling
2.9. Transcriptome Sequencing and Bioinformatics Analysis
2.10. Statistics
3. Results
3.1. Transduction Efficacy and Expression of Transgenes in UCB-MCs
3.2. Transcriptome Analysis of the Genetically Modified UCB-MCs
3.3. Secretome Profiling of the Genetically Modified UCB-MCs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aiuti, A.; Roncarolo, M.G.; Naldini, L. Gene Therapy for ADA-SCID, the First Marketing Approval of an Ex Vivo Gene Therapy in Europe: Paving the Road for the next Generation of Advanced Therapy Medicinal Products. EMBO Mol. Med. 2017, 9, 737–740. [Google Scholar] [CrossRef] [PubMed]
- Mendell, J.R.; Al-Zaidy, S.A.; Rodino-Klapac, L.R.; Goodspeed, K.; Gray, S.J.; Kay, C.N.; Boye, S.L.; Boye, S.E.; George, L.A.; Salabarria, S.; et al. Current Clinical Applications of In Vivo Gene Therapy with AAVs. Mol. Ther. 2021, 29, 464–488. [Google Scholar] [CrossRef] [PubMed]
- Sudhakar, V.; Sudhakar, R.M. Gene Therapy for Neurodegenerative Diseases. Neurotherapeutics 2019, 16, 166–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naldini, L. Ex Vivo Gene Transfer and Correction for Cell-Based Therapies. Nat. Rev. Genet. 2011, 12, 301–315. [Google Scholar] [CrossRef]
- Arjmand, B.; Larijani, B.; Sheikh Hosseini, M.; Payab, M.; Gilany, K.; Goodarzi, P.; Parhizkar Roudsari, P.; Amanollahi Baharvand, M.; Hoseini Mohammadi, N.S. The Horizon of Gene Therapy in Modern Medicine: Advances and Challenges. Adv. Exp. Med. Biol. 2020, 1247, 33–64. [Google Scholar] [CrossRef]
- Gonin, P.; Buchholz, C.J.; Pallardy, M.; Mezzina, M. Gene Therapy Bio-Safety: Scientific and Regulatory Issues. Gene Ther. 2005, 12 (Suppl. S1), S146–S152. [Google Scholar] [CrossRef] [Green Version]
- Kovacs, G.G. Molecular Pathology of Neurodegenerative Diseases: Principles and Practice. J. Clin. Pathol. 2019, 72, 725–735. [Google Scholar] [CrossRef]
- Ahuja, C.S.; Nori, S.; Tetreault, L.; Wilson, J.; Kwon, B.; Harrop, J.; Choi, D.; Fehlings, M.G. Traumatic Spinal Cord Injury—Repair and Regeneration. Neurosurgery 2017, 80, S9–S22. [Google Scholar] [CrossRef]
- Feske, S.K. Ischemic Stroke. Am. J. Med. 2021, 134, 1457–1464. [Google Scholar] [CrossRef]
- Niziolek, G.; Sandsmark, D.K.; Pascual, J.L. Neurotrauma. Curr. Opin. Crit. Care 2022, 28, 715–724. [Google Scholar] [CrossRef]
- Blaese, R.M.; Culver, K.W.; Miller, A.D.; Carter, C.S.; Fleisher, T.; Clerici, M.; Shearer, G.; Chang, L.; Chiang, Y.; Tolstoshev, P.; et al. T Lymphocyte-Directed Gene Therapy for ADA-SCID: Initial Trial Results after 4 Years. Science 1995, 270, 475–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cartier, N.; Hacein-Bey-Abina, S.; Bartholomae, C.C.; Veres, G.; Schmidt, M.; Kutschera, I.; Vidaud, M.; Abel, U.; Dal-Cortivo, L.; Caccavelli, L.; et al. Hematopoietic Stem Cell Gene Therapy with a Lentiviral Vector in X-Linked Adrenoleukodystrophy. Science 2009, 326, 818–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgan, R.A.; Gray, D.; Lomova, A.; Kohn, D.B. Hematopoietic Stem Cell Gene Therapy: Progress and Lessons Learned. Cell Stem Cell 2017, 21, 574–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bujko, K.; Kucia, M.; Ratajczak, J.; Ratajczak, M.Z. Hematopoietic Stem and Progenitor Cells (HSPCs). Adv. Exp. Med. Biol. 2019, 1201, 49–77. [Google Scholar] [CrossRef] [PubMed]
- Yu, V.W.C.; Scadden, D.T. Hematopoietic Stem Cell and Its Bone Marrow Niche. Curr. Top. Dev. Biol. 2016, 118, 21–44. [Google Scholar] [CrossRef] [PubMed]
- Orlando, N.; Pellegrino, C.; Valentini, C.G.; Bianchi, M.; Barbagallo, O.; Sparnacci, S.; Forni, F.; Fontana, T.M.; Teofili, L. Umbilical Cord Blood: Current Uses for Transfusion and Regenerative Medicine. Transfus. Apher. Sci. 2020, 59, 102952. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.A.; Luchsinger, L.L.; Greally, J.M.; Delaney, C.S. Umbilical Cord Blood: An Undervalued and Underutilized Resource in Allogeneic Hematopoietic Stem Cell Transplant and Novel Cell Therapy Applications. Curr. Opin. Hematol. 2022, 29, 317–326. [Google Scholar] [CrossRef]
- Verina, T.; Fatemi, A.; Johnston, M.V.; Comi, A.M. Pluripotent Possibilities: Human Umbilical Cord Blood Cell Treatment after Neonatal Brain Injury. Pediatr. Neurol. 2013, 48, 346–354. [Google Scholar] [CrossRef]
- Erices, A.; Conget, P.; Minguell, J.J. Mesenchymal Progenitor Cells in Human Umbilical Cord Blood. Br. J. Haematol. 2000, 109, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Ding, D.-C.; Shyu, W.-C.; Lin, S.-Z. Mesenchymal Stem Cells. Cell Transplant. 2011, 20, 5–14. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.Z.; Zhang, Y.; Wu, F.; Min, W.P.; Minev, B.; Zhang, M.; Luo, X.L.; Ramos, F.; Ichim, T.E.; Riordan, N.H.; et al. Safety Evaluation of Allogeneic Umbilical Cord Blood Mononuclear Cell Therapy for Degenerative Conditions. J. Transl. Med. 2010, 8, 75. [Google Scholar] [CrossRef] [Green Version]
- Dasari, V.R.; Spomar, D.G.; Li, L.; Gujrati, M.; Rao, J.S.; Dinh, D.H. Umbilical Cord Blood Stem Cell Mediated Downregulation of Fas Improves Functional Recovery of Rats after Spinal Cord Injury. Neurochem. Res. 2008, 33, 134–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.M.; Kurtzberg, J. Cord Blood for Brain Injury. Cytotherapy 2015, 17, 775–785. [Google Scholar] [CrossRef] [PubMed]
- Newman, M.B.; Davis, C.D.; Kuzmin-Nichols, N.; Sanberg, P.R. Human Umbilical Cord Blood (HUCB) Cells for Central Nervous System Repair. Neurotox. Res. 2003, 5, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Allan, D.S. Using Umbilical Cord Blood for Regenerative Therapy: Proof or Promise? Stem Cells 2020, 38, 590–595. [Google Scholar] [CrossRef] [PubMed]
- Newman, M.B.; Emerich, D.F.; Borlongan, C.V.; Sanberg, C.D.; Sanberg, P.R. Use of Human Umbilical Cord Blood (HUCB) Cells to Repair the Damaged Brain. Curr. Neurovasc. Res. 2004, 1, 269–281. [Google Scholar] [CrossRef]
- Fan, C.-G.; Zhang, Q.-J.; Tang, F.-W.; Han, Z.-B.; Wang, G.-S.; Han, Z.-C. Human Umbilical Cord Blood Cells Express Neurotrophic Factors. Neurosci. Lett. 2005, 380, 322–325. [Google Scholar] [CrossRef]
- Pranke, P.; Failace, R.R.; Allebrandt, W.F.; Steibel, G.; Schmidt, F.; Nardi, N.B. Hematologic and Immunophenotypic Characterization of Human Umbilical Cord Blood. Acta Haematol. 2001, 105, 71–76. [Google Scholar] [CrossRef]
- Bachstetter, A.D.; Pabon, M.M.; Cole, M.J.; Hudson, C.E.; Sanberg, P.R.; Willing, A.E.; Bickford, P.C.; Gemma, C. Peripheral Injection of Human Umbilical Cord Blood Stimulates Neurogenesis in the Aged Rat Brain. BMC Neurosci. 2008, 9, 22. [Google Scholar] [CrossRef] [Green Version]
- Ende, N.; Chen, R. Parkinson’s Disease Mice and Human Umbilical Cord Blood. J. Med. 2002, 33, 173–180. [Google Scholar]
- Garbuzova-Davis, S.; Sanberg, C.D.; Kuzmin-Nichols, N.; Willing, A.E.; Gemma, C.; Bickford, P.C.; Miller, C.; Rossi, R.; Sanberg, P.R. Human Umbilical Cord Blood Treatment in a Mouse Model of ALS: Optimization of Cell Dose. PLoS ONE 2008, 3, e2494. [Google Scholar] [CrossRef] [PubMed]
- Ende, N.; Weinstein, F.; Chen, R.; Ende, M. Human Umbilical Cord Blood Effect on Sod Mice (Amyotrophic Lateral Sclerosis). Life Sci. 2000, 67, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Knippenberg, S.; Thau, N.; Schwabe, K.; Dengler, R.; Schambach, A.; Hass, R.; Petri, S. Intraspinal Injection of Human Umbilical Cord Blood-Derived Cells Is Neuroprotective in a Transgenic Mouse Model of Amyotrophic Lateral Sclerosis. Neurodegener. Dis. 2012, 9, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Ramli, Y.; Alwahdy, A.S.; Kurniawan, M.; Juliandi, B.; Wuyung, P.E. Intra-Arterial Transplantation of Human Umbilical Cord Blood Mononuclear Cells in Sub-Acute Ischemic Stroke Increases VEGF Expression in Rats. J. Stem Cells Regen. Med. 2018, 14, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Sanberg, P.R.; Li, Y.; Wang, L.; Lu, M.; Willing, A.E.; Sanchez-Ramos, J.; Chopp, M. Intravenous Administration of Human Umbilical Cord Blood Reduces Behavioral Deficits after Stroke in Rats. Stroke 2001, 32, 2682–2688. [Google Scholar] [CrossRef] [Green Version]
- Xi, Y.; Yue, G.; Gao, S.; Ju, R.; Wang, Y. Human Umbilical Cord Blood Mononuclear Cells Transplantation for Perinatal Brain Injury. Stem Cell Res. Ther. 2022, 13, 458. [Google Scholar] [CrossRef]
- Zhu, H.; Poon, W.; Liu, Y.; Leung, G.K.-K.; Wong, Y.; Feng, Y.; Ng, S.C.P.; Tsang, K.S.; Sun, D.D.T.F.; Yeung, D.K.; et al. Phase I-II Clinical Trial Assessing Safety and Efficacy of Umbilical Cord Blood Mononuclear Cell Transplant Therapy of Chronic Complete Spinal Cord Injury. Cell Transplant. 2016, 25, 1925–1943. [Google Scholar] [CrossRef] [Green Version]
- Islamov, R.R.; Rizvanov, A.A.; Mukhamedyarov, M.A.; Salafutdinov, I.I.; Garanina, E.E.; Fedotova, V.Y.; Solovyeva, V.V.; Mukhamedshina, Y.O.; Safiullov, Z.Z.; Izmailov, A.A.; et al. Symptomatic Improvement, Increased Life-Span and Sustained Cell Homing in Amyotrophic Lateral Sclerosis after Transplantation of Human Umbilical Cord Blood Cells Genetically Modified with Adeno-Viral Vectors Expressing a Neuro-Protective Factor and a Neur. Curr. Gene Ther. 2015, 15, 266–276. [Google Scholar] [CrossRef]
- Sathasivam, S. VEGF and ALS. Neurosci. Res. 2008, 62, 71–77. [Google Scholar] [CrossRef]
- Lange, C.; Storkebaum, E.; De Almodóvar, C.R.; Dewerchin, M.; Carmeliet, P. Vascular Endothelial Growth Factor: A Neurovascular Target in Neurological Diseases. Nat. Rev. Neurol. 2016, 12, 439–454. [Google Scholar] [CrossRef]
- Ferrara, N.; Gerber, H.-P.; LeCouter, J. The Biology of VEGF and Its Receptors. Nat. Med. 2003, 9, 669–676. [Google Scholar] [CrossRef] [PubMed]
- De Almodovar, C.R.; Lambrechts, D.; Mazzone, M.; Carmeliet, P.; Ruiz de Almodovar, C.; Lambrechts, D.; Mazzone, M.; Carmeliet, P. Role and Therapeutic Potential of VEGF in the Nervous System. Physiol. Rev. 2009, 89, 607–648. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zechariah, A.; Qu, Y.; Hermann, D.M. Effects of Vascular Endothelial Growth Factor in Ischemic Stroke. J. Neurosci. Res. 2012, 90, 1873–1882. [Google Scholar] [CrossRef]
- Chen, B.; Zhang, Y.; Chen, S.; Xuran, L.; Dong, J.; Chen, W.; Tao, S.; Yang, W.; Zhang, Y. The Role of Vascular Endothelial Growth Factor in Ischemic Stroke. Pharmazie 2021, 76, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Gao, S.; Li, Y.; Peng, R.; Zheng, Z.; Wei, W.; Zhao, Z.; Liu, X.; Li, L.; Zhang, J. VEGI Improves Outcomes in the Early Phase of Experimental Traumatic Brain Injury. Neuroscience 2020, 438, 60–69. [Google Scholar] [CrossRef]
- Thau-Zuchman, O.; Shohami, E.; Alexandrovich, A.G.; Leker, R.R. Vascular Endothelial Growth Factor Increases Neurogenesis after Traumatic Brain Injury. J. Cereb. Blood Flow Metab. 2010, 30, 1008–1016. [Google Scholar] [CrossRef] [Green Version]
- Siddiq, I.; Park, E.; Liu, E.; Spratt, S.K.; Surosky, R.; Lee, G.; Ando, D.; Giedlin, M.; Hare, G.M.T.; Fehlings, M.G.; et al. Treatment of Traumatic Brain Injury Using Zinc-Finger Protein Gene Therapy Targeting VEGF-A. J. Neurotrauma 2012, 29, 2647–2659. [Google Scholar] [CrossRef]
- Li, J.; Chen, S.; Zhao, Z.; Luo, Y.; Hou, Y.; Li, H.; He, L.; Zhou, L.; Wu, W. Effect of VEGF on Inflammatory Regulation, Neural Survival, and Functional Improvement in Rats Following a Complete Spinal Cord Transection. Front. Cell. Neurosci. 2017, 11, 381. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Xu, W.; Zhang, Z.; Liu, H.; Lv, L.; Han, D.; Liu, L.; Yao, A.; Xu, T. Vascular Endothelial Growth Factor-Transfected Bone Marrow Mesenchymal Stem Cells Improve the Recovery of Motor and Sensory Functions of Rats With Spinal Cord Injury. Spine 2020, 45, E364–E372. [Google Scholar] [CrossRef]
- Pronto-Laborinho, A.C.; Pinto, S.; de Carvalho, M. Roles of Vascular Endothelial Growth Factor in Amyotrophic Lateral Sclerosis. Biomed. Res. Int. 2014, 2014, 947513. [Google Scholar] [CrossRef] [Green Version]
- Lambrechts, D.; Storkebaum, E.; Morimoto, M.; Del-Favero, J.; Desmet, F.; Marklund, S.L.; Wyns, S.; Thijs, V.; Andersson, J.; van Marion, I.; et al. VEGF Is a Modifier of Amyotrophic Lateral Sclerosis in Mice and Humans and Protects Motoneurons against Ischemic Death. Nat. Genet. 2003, 34, 383–394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agatieva, E.; Ksembaev, S.; Sokolov, M.; Markosyan, V.; Gazizov, I.; Tsyplakov, D.; Shmarov, M.; Tutykhina, I.; Naroditsky, B.; Logunov, D.; et al. Evaluation of Direct and Cell-Mediated Lactoferrin Gene Therapy for the Maxillofacial Area Abscesses in Rats. Pharmaceutics 2021, 13, 58. [Google Scholar] [CrossRef] [PubMed]
- Gatina, D.Z.; Gazizov, I.M.; Zhuravleva, M.N.; Arkhipova, S.S.; Golubenko, M.A.; Gomzikova, M.O.; Garanina, E.E.; Islamov, R.R.; Rizvanov, A.A.; Salafutdinov, I.I. Induction of Angiogenesis by Genetically Modified Human Umbilical Cord Blood Mononuclear Cells. Int. J. Mol. Sci. 2023, 24, 4396. [Google Scholar] [CrossRef] [PubMed]
- Islamov, R.R.; Izmailov, A.A.; Sokolov, M.E.; Fadeev, F.O.; Bashirov, F.V.; Eremeev, A.A.; Shmarov, M.M.; Naroditskiy, B.S.; Chelyshev, Y.A.A.; Lavrov, I.A.; et al. Evaluation of Direct and Cell-Mediated Triple-Gene Therapy in Spinal Cord Injury in Rats. Brain Res. Bull. 2017, 132, 44–52. [Google Scholar] [CrossRef]
- Markosyan, V.; Safiullov, Z.; Izmailov, A.; Fadeev, F.; Sokolov, M.; Kuznetsov, M.; Trofimov, D.; Kim, E.; Kundakchyan, G.; Gibadullin, A.; et al. Preventive Triple Gene Therapy Reduces the Negative Consequences of Ischemia-Induced Brain Injury after Modelling Stroke in a Rat. Int. J. Mol. Sci. 2020, 21, 6858. [Google Scholar] [CrossRef]
- Baer, A.; Kehn-Hall, K. Viral Concentration Determination through Plaque Assays: Using Traditional and Novel Overlay Systems. J. Vis. Exp. 2014, 4, e52065. [Google Scholar] [CrossRef]
- Chu, Y.W.; Wang, R.; Schmid, I.; Sakamoto, K.M. Analysis with Flow Cytometry of Green Fluorescent Protein Expression in Leukemic Cells. Cytometry 1999, 36, 333–339. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Garanina, E.E.; Mukhamedshina, Y.O.; Salafutdinov, I.I.; Kiyasov, A.P.; Lima, L.M.; Reis, H.J.; Palotás, A.; Islamov, R.R.; Rizvanov, A.A. Construction of Recombinant Adenovirus Containing Picorna-Viral 2A-Peptide Sequence for the Co-Expression of Neuro-Protective Growth Factors in Human Umbilical Cord Blood Cells. Spinal Cord 2016, 54, 423–430. [Google Scholar] [CrossRef] [Green Version]
- Clark, M.F.; Lister, R.M.; Bar-Joseph, M. ELISA Techniques. In Plant Molecular Biology; Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1986; Volume 118, pp. 742–766. [Google Scholar]
- Valekova, I.; Skalnikova, H.K.; Jarkovska, K.; Motlik, J.; Kovarova, H. Multiplex Immunoassays for Quantification of Cytokines, Growth Factors, and Other Proteins in Stem Cell Communication. Methods Mol. Biol. 2015, 1212, 39–63. [Google Scholar] [CrossRef]
- Kukurba, K.R.; Montgomery, S.B. RNA Sequencing and Analysis. Cold Spring Harb. Protoc. 2015, 2015, 951–969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-Optimal Probabilistic RNA-Seq Quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene Set Analysis Toolkit with Revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steffin, D.H.M.; Hsieh, E.M.; Rouce, R.H. Gene Therapy: Current Applications and Future Possibilities. Adv. Pediatr. 2019, 66, 37–54. [Google Scholar] [CrossRef]
- Blind, J.E.; McLeod, E.N.; Brown, A.; Patel, H.; Ghosh, S. Biosafety Practices for In Vivo Viral-Mediated Gene Therapy in the Health Care Setting. Appl. Biosaf. 2020, 25, 194–200. [Google Scholar] [CrossRef]
- High, K.A.; Roncarolo, M.G. Gene Therapy. N. Engl. J. Med. 2019, 381, 455–464. [Google Scholar] [CrossRef]
- Berglund, S.; Magalhaes, I.; Gaballa, A.; Vanherberghen, B.; Uhlin, M. Advances in Umbilical Cord Blood Cell Therapy: The Present and the Future. Expert Opin. Biol. Ther. 2017, 17, 691–699. [Google Scholar] [CrossRef]
- Ikeda, Y.; Fukuda, N.; Wada, M.; Matsumoto, T.; Satomi, A.; Yokoyama, S.-I.; Saito, S.; Matsumoto, K.; Kanmatsuse, K.; Mugishima, H. Development of Angiogenic Cell and Gene Therapy by Transplantation of Umbilical Cord Blood with Vascular Endothelial Growth Factor Gene. Hypertens. Res. 2004, 27, 119–128. [Google Scholar] [CrossRef] [Green Version]
- Das, H.; George, J.C.; Joseph, M.; Das, M.; Abdulhameed, N.; Blitz, A.; Khan, M.; Sakthivel, R.; Mao, H.-Q.; Hoit, B.D.; et al. Stem Cell Therapy with Overexpressed VEGF and PDGF Genes Improves Cardiac Function in a Rat Infarct Model. PLoS ONE 2009, 4, e7325. [Google Scholar] [CrossRef]
- Chen, H.K.; Hung, H.F.; Shyu, K.G.; Wang, B.W.; Sheu, J.R.; Liang, Y.J.; Chang, C.C.; Kuan, P. Combined Cord Blood Stem Cells and Gene Therapy Enhances Angiogenesis and Improves Cardiac Performance in Mouse after Acute Myocardial Infarction. Eur. J. Clin. Investig. 2005, 35, 677–686. [Google Scholar] [CrossRef]
- Islamov, R.R.; Rizvanov, A.A.; Fedotova, V.Y.; Izmailov, A.A.; Safiullov, Z.Z.; Garanina, E.E.; Salafutdinov, I.I.; Sokolov, M.E.; Mukhamedyarov, M.A.; Palotás, A. Tandem Delivery of Multiple Therapeutic Genes Using Umbilical Cord Blood Cells Improves Symptomatic Outcomes in ALS. Mol. Neurobiol. 2017, 54, 4756–4763. [Google Scholar] [CrossRef] [PubMed]
- Islamov, R.; Bashirov, F.; Fadeev, F.; Shevchenko, R.; Izmailov, A.; Markosyan, V.; Sokolov, M.; Kuznetsov, M.; Davleeva, M.; Garifulin, R.; et al. Epidural Stimulation Combined with Triple Gene Therapy for Spinal Cord Injury Treatment. Int. J. Mol. Sci. 2020, 21, 8896. [Google Scholar] [CrossRef] [PubMed]






| Name | Nucleotide Sequence |
|---|---|
| β-actin-TM-Forward (human) | GCGAGAAGATGACCCAGGATC |
| β-actin-TM-Reverse (human) | CCAGTGGTACGGCCAGAGG |
| β-actin-TM-Probe (human) | [FAM]CCAGCCATGTACGTTGCTATCCAGGC[BH1] |
| hVEGF-TM49-Forward (human) | TACCTCCACCATGCCAAGTG |
| hVEGF-TM110-Reverse (human) | TGATTCTGCCCTCCTCCTTCT |
| hVEGF-TM-Probe (human) | [FAM]TCCCAGGCTGCACCCATGG[BH1] |
| GFP-TM-Forward | AGCAAAGACCCCAACGAGAA |
| GFP-TM-Reverse | GGCGGCGGTCACGAA |
| GFP-TM-Probe | [FAM]CGCGATCACATGGTCCTGCTGG[BH1] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salafutdinov, I.I.; Gatina, D.Z.; Markelova, M.I.; Garanina, E.E.; Malanin, S.Y.; Gazizov, I.M.; Izmailov, A.A.; Rizvanov, A.A.; Islamov, R.R.; Palotás, A.; et al. A Biosafety Study of Human Umbilical Cord Blood Mononuclear Cells Transduced with Adenoviral Vector Carrying Human Vascular Endothelial Growth Factor cDNA In Vitro. Biomedicines 2023, 11, 2020. https://doi.org/10.3390/biomedicines11072020
Salafutdinov II, Gatina DZ, Markelova MI, Garanina EE, Malanin SY, Gazizov IM, Izmailov AA, Rizvanov AA, Islamov RR, Palotás A, et al. A Biosafety Study of Human Umbilical Cord Blood Mononuclear Cells Transduced with Adenoviral Vector Carrying Human Vascular Endothelial Growth Factor cDNA In Vitro. Biomedicines. 2023; 11(7):2020. https://doi.org/10.3390/biomedicines11072020
Chicago/Turabian StyleSalafutdinov, Ilnur I., Dilara Z. Gatina, Maria I. Markelova, Ekaterina E. Garanina, Sergey Yu. Malanin, Ilnaz M. Gazizov, Andrei A. Izmailov, Albert A. Rizvanov, Rustem R. Islamov, András Palotás, and et al. 2023. "A Biosafety Study of Human Umbilical Cord Blood Mononuclear Cells Transduced with Adenoviral Vector Carrying Human Vascular Endothelial Growth Factor cDNA In Vitro" Biomedicines 11, no. 7: 2020. https://doi.org/10.3390/biomedicines11072020
APA StyleSalafutdinov, I. I., Gatina, D. Z., Markelova, M. I., Garanina, E. E., Malanin, S. Y., Gazizov, I. M., Izmailov, A. A., Rizvanov, A. A., Islamov, R. R., Palotás, A., & Safiullov, Z. Z. (2023). A Biosafety Study of Human Umbilical Cord Blood Mononuclear Cells Transduced with Adenoviral Vector Carrying Human Vascular Endothelial Growth Factor cDNA In Vitro. Biomedicines, 11(7), 2020. https://doi.org/10.3390/biomedicines11072020