A Human DUB Protein Array for Clarification of Linkage Specificity of Polyubiquitin Chain and Application to Evaluation of Its Inhibitors
Abstract
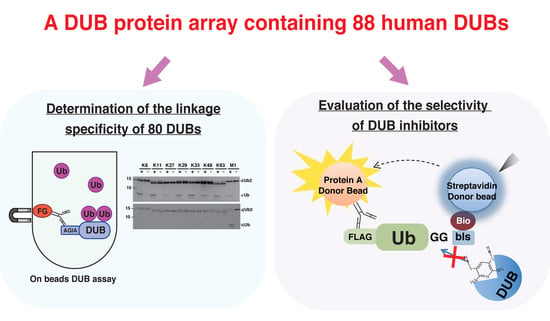
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Construction of Expression Vectors Containing Human DUB cDNAs
2.3. Synthesis of Recombinant DUB Proteins
2.4. Preparation of Anti-AGIA Antibody-Conjugated Magnetic Beads
2.5. In Vitro Deubiquitination Assay
2.6. Inhibitor Assay Using AlphaScreen
3. Results
3.1. Preparation of Human DUB Protein Array
3.2. Establishment of an In Vitro DUB Assay
3.3. Comprehensive DUB Assay Using the DUB Protein Array
3.4. Application of the DUB Array for the Evaluation of DUB Inhibitor Selectivity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| DUB | Deubiquitinating enzyme |
| USP | Ubiquitin specific protease |
| OTU | Ovarian tumor |
| UCH | Ubiquitin C-terminal hydrolase |
| JAMM | JAB1/MPN/Mov34 |
| CBB | Coomassie Brilliant Blue |
References
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nijman, S.M.B.; Luna-Vargas, M.P.A.; Velds, A.; Brummelkamp, T.R.; Dirac, A.M.G.; Sixma, T.K.; Bernards, R. A genomic and functional inventory of deubiquitinating enzymes. Cell 2005, 123, 773–786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singhal, S.; Taylor, M.C.; Baker, R.T. Deubiquitylating enzymes and disease. BMC Biochem. 2008, 9 (Suppl. 1), S3. [Google Scholar] [CrossRef] [Green Version]
- Hussain, S.; Zhang, Y.; Galardy, P.J. DUBs and cancer: The role of deubiquitinating enzymes as oncogenes, non-oncogenes and tumor suppressors. Cell Cycle 2009, 8, 1688–1697. [Google Scholar] [CrossRef]
- Wei, R.; Liu, X.; Yu, W.; Yang, T.; Cai, W.; Liu, J.; Huang, X.; Xu, G.-T.; Zhao, S.; Yang, J.; et al. Deubiquitinases in cancer. Oncotarget 2015, 6, 12872–12889. [Google Scholar] [CrossRef] [Green Version]
- Wertz, I.E.; Wang, X. From discovery to bedside: Targeting the ubiquitin system. Cell Chem Biol. 2019, 26, 156–177. [Google Scholar] [CrossRef]
- Colland, F.; Formstecher, E.; Jacq, X.; Reverdy, C.; Planquette, C.; Conrath, S.; Trouplin, V.; Bianchi, J.; Aushev, V.N.; Camonis, J.; et al. Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol. Cancer Ther. 2009, 8, 2286–2295. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.-H.; Lee, M.J.; Park, S.; Oh, D.-C.; Elsasser, S.; Chen, P.-C.; Gartner, C.; Dimova, N.; Hanna, J.; Gygi, S.P.; et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 2010, 467, 179–184. [Google Scholar] [CrossRef] [Green Version]
- Eletr, Z.M.; Wilkinson, K.D. Regulation of proteolysis by human deubiquitinating enzymes. Biochim. Biophys. Acta 2014, 1843, 114–128. [Google Scholar] [CrossRef] [Green Version]
- Abdul Rehman, S.A.; Kristariyanto, Y.A.; Choi, S.-Y.; Nkosi, P.J.; Weidlich, S.; Labib, K.; Hofmann, K.; Kulathu, Y. MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol. Cell 2016, 63, 146–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hermanns, T.; Pichlo, C.; Woiwode, I.; Klopffleisch, K.; Witting, K.F.; Ovaa, H.; Baumann, U.; Hofmann, K. A family of unconventional deubiquitinases with modular chain specificity determinants. Nat. Commun. 2018, 9, 799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hewings, D.S.; Heideker, J.; Ma, T.P.; AhYoung, A.P.; El Oualid, F.; Amore, A.; Costakes, G.T.; Kirchhofer, D.; Brasher, B.; Pillow, T.; et al. Reactive-site-centric chemoproteomics identifies a distinct class of deubiquitinase enzymes. Nat. Commun. 2018, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Amerik, A.Y.; Hochstrasser, M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta 2004, 1695, 189–207. [Google Scholar] [CrossRef] [Green Version]
- Komander, D.; Lord, C.J.; Scheel, H.; Swift, S.; Hofmann, K.; Ashworth, A.; Barford, D. The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol. Cell 2008, 29, 451–464. [Google Scholar] [CrossRef]
- Komander, D.; Reyes-Turcu, F.; Licchesi, J.D.F.; Odenwaelder, P.; Wilkinson, K.D.; Barford, D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009, 10, 466–473. [Google Scholar] [CrossRef] [Green Version]
- Sato, Y.; Goto, E.; Shibata, Y.; Kubota, Y.; Yamagata, A.; Goto-Ito, S.; Kubota, K.; Inoue, J.-I.; Takekawa, M.; Tokunaga, F.; et al. Structures of CYLD USP with Met1- or Lys63-linked diubiquitin reveal mechanisms for dual specificity. Nat. Struct. Mol. Biol. 2015, 22, 222–229. [Google Scholar] [CrossRef]
- Mevissen, T.E.T.; Hospenthal, M.K.; Geurink, P.P.; Elliott, P.R.; Akutsu, M.; Arnaudo, N.; Ekkebus, R.; Kulathu, Y.; Wauer, T.; El Oualid, F.; et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell 2013, 154, 169–184. [Google Scholar] [CrossRef] [Green Version]
- Mevissen, T.E.T.; Kulathu, Y.; Mulder, M.P.C.; Geurink, P.P.; Maslen, S.L.; Gersch, M.; Elliott, P.R.; Burke, J.E.; van Tol, B.D.M.; Akutsu, M.; et al. Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne. Nature 2016, 538, 402–405. [Google Scholar] [CrossRef]
- Faesen, A.C.; Luna-Vargas, M.P.A.; Geurink, P.P.; Clerici, M.; Merkx, R.; van Dijk, W.J.; Hameed, D.S.; Hameed, D.S.; Hameed, D.S.; El Oualid, F.; et al. The differential modulation of USP activity by internal regulatory domains, interactors and eight ubiquitin chain types. Chem. Biol. 2011, 18, 1550–1561. [Google Scholar] [CrossRef] [Green Version]
- Ritorto, M.S.; Ewan, R.; Perez-Oliva, A.B.; Knebel, A.; Buhrlage, S.J.; Wightman, M.; Kelly, S.M.; Wood, N.T.; Virdee, S.; Gray, N.S.; et al. Screening of DUB activity and specificity by MALDI-TOF mass spectrometry. Nat. Commun. 2014, 5, 4763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.-H.; Zhou, C.-J.; Zhou, Z.-R.; Song, A.-X.; Hu, H.-Y. Domain Analysis Reveals That a Deubiquitinating Enzyme USP13 Performs Non-Activating Catalysis for Lys63-Linked Polyubiquitin. PLoS ONE 2011, 6, e29362-13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winborn, B.J.; Travis, S.M.; Todi, S.V.; Scaglione, K.M.; Xu, P.; Williams, A.J.; Cohen, R.E.; Peng, J.; Paulson, H.L. The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits Lys63 linkages in mixed linkage ubiquitin chains. J. Biol. Chem. 2008, 283, 26436–26443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamanaka, S.; Sato, Y.; Oikawa, D.; Goto, E.; Fukai, S.; Tokunaga, F.; Takahashi, H.; Sawasaki, T. Subquinocin, a small molecule inhibitor of CYLD and USP-family deubiquitinating enzymes, promotes NF-κB signaling. Biochem. Biophys. Res. Commun. 2020, 524, 1–7. [Google Scholar] [CrossRef]
- Takahashi, H.; Uematsu, A.; Yamanaka, S.; Imamura, M.; Nakajima, T.; Doi, K.; Yasuoka, S.; Takahashi, C.; Takeda, H.; Sawasaki, T. Establishment of a wheat cell-free synthesized protein array containing 250 human and mouse E3 ubiquitin ligases to identify novel interaction between E3 ligases and substrate proteins. PLoS ONE 2016, 11, e0156718. [Google Scholar] [CrossRef] [Green Version]
- Yano, T.; Takeda, H.; Uematsu, A.; Yamanaka, S.; Nomura, S.; Nemoto, K.; Iwasaki, T.; Takahashi, H.; Sawasaki, T. AGIA Tag System Based on a High Affinity Rabbit Monoclonal Antibody against Human Dopamine Receptor D1 for Protein Analysis. PLoS ONE 2016, 11, e0156716. [Google Scholar] [CrossRef]
- McGouran, J.F.; Gaertner, S.R.; Altun, M.; Kramer, H.B.; Kessler, B.M. Deubiquitinating enzyme specificity for ubiquitin chain topology profiled by di-ubiquitin activity probes. Chem. Biol. 2013, 20, 1447–1455. [Google Scholar] [CrossRef] [Green Version]
- Ceccarelli, D.F.; Ivantsiv, S.; Mullin, A.A.; Coyaud, E.; Manczyk, N.; Maisonneuve, P.; Kurinov, I.; Zhao, L.; Go, C.; Gingras, A.-C.; et al. FAM105A/OTULINL is a pseudodeubiquitinase of the OTU-class that localizes to the ER membrane. Structure 2019, 27, 1000–1012.e6. [Google Scholar] [CrossRef]
- Weeks, S.D.; Grasty, K.C.; Hernandez-Cuebas, L.; Loll, P.J. Crystal structure of a Josephin-ubiquitin complex: Evolutionary restraints on ataxin-3 deubiquitinating activity. J. Biol. Chem. 2011, 286, 4555–4565. [Google Scholar] [CrossRef] [Green Version]
- Yi, F.; Regan, L. A Novel Class of Small Molecule Inhibitors of Hsp90. ACS Chem. Biol. 2008, 3, 645–654. [Google Scholar] [CrossRef] [Green Version]
- Uematsu, A.; Kido, K.; Manabe, E.; Takeda, H.; Takahashi, H.; Hayashi, M.; Imai, Y.; Sawasaki, T. DANFIN functions as an inhibitor of transcription factor NF-κB and potentiates the antitumor effect of bortezomib in multiple myeloma. Biochem. Biophys. Res. Commun. 2018, 495, 2289–2295. [Google Scholar] [CrossRef] [PubMed]
- Nomura, S.; Takahashi, H.; Suzuki, J.; Kuwahara, M.; Yamashita, M.; Sawasaki, T. Pyrrothiogatain acts as an inhibitor of GATA family proteins and inhibits Th2 cell differentiation in vitro. Sci. Rep. 2019, 9, 17335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, H.; Takahashi, C.; Moreland, N.J.; Chang, Y.-T.; Sawasaki, T.; Ryo, A.; Vasudevan, S.G.; Suzuki, Y.; Yamamoto, N. Establishment of a robust dengue virus NS3-NS5 binding assay for identification of protein-protein interaction inhibitors. Antiviral Res. 2012, 96, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Schulz, S.; Chachami, G.; Kozaczkiewicz, L.; Winter, U.; Stankovic-Valentin, N.; Haas, P.; Hofmann, K.; Urlaub, H.; Ovaa, H.; Wittbrodt, J.; et al. Ubiquitin-specific protease-like 1 (USPL1) is a SUMO isopeptidase with essential, non-catalytic functions. EMBO Rep. 2012, 13, 930–938. [Google Scholar] [CrossRef] [Green Version]
- Cohn, M.A.; Kowal, P.; Yang, K.; Haas, W.; Huang, T.T.; Gygi, S.P.; D’Andrea, A.D. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol. Cell 2007, 28, 786–797. [Google Scholar] [CrossRef]
- Sato, Y.; Yoshikawa, A.; Yamagata, A.; Mimura, H.; Yamashita, M.; Ookata, K.; Nureki, O.; Iwai, K.; Komada, M.; Fukai, S. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature 2008, 455, 358–362. [Google Scholar] [CrossRef]
- Yao, T.; Song, L.; Xu, W.; DeMartino, G.N.; Florens, L.; Swanson, S.K.; Washburn, M.P.; Conaway, R.C.; Conaway, J.W.; Cohen, R.E. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat. Cell Biol. 2006, 8, 994–1002. [Google Scholar] [CrossRef]
- Altun, M.; Kramer, H.B.; Willems, L.I.; McDermott, J.L.; Leach, C.A.; Goldenberg, S.J.; Kumar, K.G.S.; Konietzny, R.; Fischer, R.; Kogan, E.; et al. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem. Biol. 2011, 18, 1401–1412. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Dexheimer, T.S.; Ai, Y.; Liang, Q.; Villamil, M.A.; Inglese, J.; Maloney, D.J.; Jadhav, A.; Simeonov, A.; Zhuang, Z. Selective and cell-active inhibitors of the USP1/UAF1 deubiquitinase complex reverse cisplatin resistance in non-small cell lung cancer cells. Chem. Biol. 2011, 18, 1390–1400. [Google Scholar] [CrossRef] [Green Version]
- Mistry, H.; Hsieh, G.; Buhrlage, S.J.; Huang, M.; Park, E.; Cuny, G.D.; Galinsky, I.; Stone, R.M.; Gray, N.S.; D’Andrea, A.D.; et al. Small-molecule inhibitors of USP1 target ID1 degradation in leukemic cells. Mol. Cancer Ther. 2013, 12, 2651–2662. [Google Scholar] [CrossRef] [Green Version]
- Ndubaku, C.; Tsui, V. Inhibiting the deubiquitinating enzymes (DUBs). J. Med. Chem. 2015, 58, 1581–1595. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, A.P.; Ioannidis, S.; Krajewski, W.W.; Pinto-Fernandez, A.; Heride, C.; Martin, A.C.L.; Tonkin, L.M.; Townsend, E.C.; Buker, S.M.; Lancia, D.R.; et al. Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature 2017, 550, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Kategaya, L.; Di Lello, P.; Rougé, L.; Pastor, R.; Clark, K.R.; Drummond, J.; Kleinheinz, T.; Lin, E.; Upton, J.-P.; Prakash, S.; et al. USP7 small-molecule inhibitors interfere with ubiquitin binding. Nature 2017, 550, 534–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Family (in Human Genome) | Expression Construct | Successfully Synthesized | Active DUBs | Inactive DUBs |
|---|---|---|---|---|
| USP (56) | 54 | 54 | 52 | 2 |
| OTU (17) | 16 | 16 | 14 | 2 |
| UCH (4) | 4 | 4 | 2 | 2 |
| Josephin (4) | 4 | 4 | 4 | 0 |
| JAMM (12) | 11 | 10 | 8 | 2 |
| Total (93) | 89 | 88 | 80 | 8 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takahashi, H.; Yamanaka, S.; Kuwada, S.; Higaki, K.; Kido, K.; Sato, Y.; Fukai, S.; Tokunaga, F.; Sawasaki, T. A Human DUB Protein Array for Clarification of Linkage Specificity of Polyubiquitin Chain and Application to Evaluation of Its Inhibitors. Biomedicines 2020, 8, 152. https://doi.org/10.3390/biomedicines8060152
Takahashi H, Yamanaka S, Kuwada S, Higaki K, Kido K, Sato Y, Fukai S, Tokunaga F, Sawasaki T. A Human DUB Protein Array for Clarification of Linkage Specificity of Polyubiquitin Chain and Application to Evaluation of Its Inhibitors. Biomedicines. 2020; 8(6):152. https://doi.org/10.3390/biomedicines8060152
Chicago/Turabian StyleTakahashi, Hirotaka, Satoshi Yamanaka, Shohei Kuwada, Kana Higaki, Kohki Kido, Yusuke Sato, Shuya Fukai, Fuminori Tokunaga, and Tatsuya Sawasaki. 2020. "A Human DUB Protein Array for Clarification of Linkage Specificity of Polyubiquitin Chain and Application to Evaluation of Its Inhibitors" Biomedicines 8, no. 6: 152. https://doi.org/10.3390/biomedicines8060152
APA StyleTakahashi, H., Yamanaka, S., Kuwada, S., Higaki, K., Kido, K., Sato, Y., Fukai, S., Tokunaga, F., & Sawasaki, T. (2020). A Human DUB Protein Array for Clarification of Linkage Specificity of Polyubiquitin Chain and Application to Evaluation of Its Inhibitors. Biomedicines, 8(6), 152. https://doi.org/10.3390/biomedicines8060152