SARS-CoV-2 RdRp Inhibitors Selected from a Cell-Based SARS-CoV-2 RdRp Activity Assay System
Abstract
:1. Introduction
2. Materials and Methods
2.1. Test Compounds
2.2. Plasmid Construction
2.3. Cells and Transfection
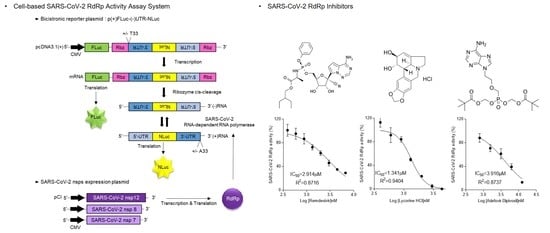
2.4. Cell-Based SARS-CoV-2 RdRp Activity Assay
2.5. Western Blot Assay
2.6. Immunofluorescence Staining Assay
2.7. Calculation of Z-Factor
2.8. Statistical Analysis
3. Results
3.1. Establishment of the Cell-Based SARS-CoV-2 RdRp Activity Assay System
3.2. Effect of Accessory Proteins nsp7 and nsp8 SARS-CoV-2 RdRp Activity
3.3. Validation of the Cell-Based SARS-CoV-2 RdRp Activity Assay System
3.4. Inhibition of SARS-CoV-2 RdRp Activity by Remdesivir and Lycorine
3.5. Inhibition of SARS-CoV RdRp Activity by Nucleos(t)ide
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 3CLpro | 3C-like protease |
| CoV | coronavirus |
| COVID-19 | coronavirus disease 2019 |
| C-term | C-terminal |
| FDA | Food and Drug Administration |
| FLuc | firefly luciferase |
| HBV | hepatitis B virus |
| HDV | hepatitis delta virus |
| HIV | human immunodeficiency virus |
| HSV | herpes simplex virus |
| HTS | high-throughput screening |
| IRES | internal ribosome entry site |
| MERS | Middle East respiratory syndrome |
| NLuc | Nano-glo® luciferase |
| NSP | Nonstructural proteins |
| NTP | Nucleoside triphosphate |
| N-term | N-terminal |
| ORF | open reading frame |
| PLpro | papain-like protease |
| RdRp | RNA-dependent RNA polymerase |
| SARS | severe acute respiratory syndrome |
| SARS-CoV-2 | severe acute respiratory syndrome coronavirus 2 |
| UTR | untranslated region |
References
- World Health Organization. Coronavirus Disease (COVID-19)—World Health Organization. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 12 March 2021).
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Lee, J.-Y.; Yang, J.-S.; Kim, J.W.; Kim, V.N.; Chang, H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020, 181, 914–921.e10. [Google Scholar] [CrossRef]
- V’Kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2020, 19, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, T.; Sun, Q.; Ming, Z.; Zhang, L.; et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020, 368, 779–782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Picarazzi, F.; Vicenti, I.; Saladini, F.; Zazzi, M.; Mori, M. Targeting the RdRp of Emerging RNA Viruses: The Structure-Based Drug Design Challenge. Molecules 2020, 25, 5695. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, P.; Niyogi, S. SARS-CoV-2 mutations: The biological trackway towards viral fitness. Epidemiol. Infect. 2021, 149, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Mao, C.; Luan, X.; Shen, D.-D.; Shen, Q.; Su, H.; Wang, X.; Zhou, F.; Zhao, W.; Gao, M.; et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 2020, 368, 1499–1504. [Google Scholar] [CrossRef]
- Gordon, C.J.; Tchesnokov, E.P.; Feng, J.Y.; Porter, D.P.; Götte, M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020, 295, 4773–4779. [Google Scholar] [CrossRef] [Green Version]
- Velthuis, A.T.; Arnold, J.J.; Cameron, C.E.; Worm, S.H.E.V.D.; Snijder, E.J. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 2009, 38, 203–214. [Google Scholar] [CrossRef] [Green Version]
- Min, J.S.; Kim, G.-W.; Kwon, S.; Jin, Y.-H. A Cell-Based Reporter Assay for Screening Inhibitors of MERS Coronavirus RNA-Dependent RNA Polymerase Activity. J. Clin. Med. 2020, 9, 2399. [Google Scholar] [CrossRef]
- Sui, Y.; Wu, Z. Alternative Statistical Parameter for High-Throughput Screening Assay Quality Assessment. J. Biomol. Screen. 2007, 12, 229–234. [Google Scholar] [CrossRef] [Green Version]
- Dreyfus, M.; Régnier, P. The Poly(A) Tail of mRNAs: Bodyguard in Eukaryotes, Scavenger in Bacteria. Cell 2002, 111, 611–613. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Wu, J.; Wang, H.; Gao, Y.; Liu, Q.; Mu, A.; Ji, W.; Yan, L.; Zhu, Y.; Zhu, C.; et al. Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase. Cell 2020, 182, 417–428.e13. [Google Scholar] [CrossRef] [PubMed]
- Kirchdoerfer, R.N.; Ward, A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019, 10, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Subissi, L.; Posthuma, C.C.; Collet, A.; Zevenhoven-Dobbe, J.C.; Gorbalenya, A.; Decroly, E.; Snijder, E.; Canard, B.; Imbert, I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. USA 2014, 111, E3900–E3909. [Google Scholar] [CrossRef] [Green Version]
- Hillen, H.S.; Kokic, G.; Farnung, L.; Dienemann, C.; Tegunov, D.; Cramer, P. Structure of replicating SARS-CoV-2 polymerase. Nature 2020, 584, 154–156. [Google Scholar] [CrossRef]
- Beigel, J.H.; Tomashek, K.M.; Dodd, L.E.; Mehta, A.K.; Zingman, B.S.; Kalil, A.C.; Hohmann, E.; Chu, H.Y.; Luetkemeyer, A.; Kline, S.; et al. Remdesivir for the Treatment of Covid-19—Final Report. N. Engl. J. Med. 2020, 383, 1813–1826. [Google Scholar] [CrossRef] [PubMed]
- Agostini, M.L.; Andres, E.L.; Sims, A.C.; Graham, R.L.; Sheahan, T.P.; Lu, X.; Smith, E.C.; Case, J.B.; Feng, J.Y.; Jordan, R.; et al. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. mBio 2018, 9, e00221-18. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.-H.; Min, J.S.; Jeon, S.; Lee, J.; Kim, S.; Park, T.; Park, D.; Jang, M.S.; Park, C.M.; Song, J.H.; et al. Lycorine, a non-nucleoside RNA dependent RNA polymerase inhibitor, as potential treatment for emerging coronavirus infections. Phytomedicine 2021, 86, 153440. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.E.; Min, J.S.; Jang, M.S.; Lee, J.Y.; Shin, Y.S.; Park, C.M.; Song, J.H.; Kim, H.R.; Kim, S.; Jin, Y.-H.; et al. Natural Bis-Benzylisoquinoline Alkaloids-Tetrandrine, Fangchinoline, and Cepharanthine, Inhibit Human Coronavirus OC43 Infection of MRC-5 Human Lung Cells. Biomolecules 2019, 9, 696. [Google Scholar] [CrossRef] [Green Version]
- Ohashi, H.; Watashi, K.; Saso, W.; Shionoya, K.; Iwanami, S.; Hirokawa, T.; Shirai, T.; Kanaya, S.; Ito, Y.; Kim, K.S.; et al. Potential anti-COVID-19 agents, cepharanthine and nelfinavir, and their usage for combination treatment. iScience 2021, 24, 102367. [Google Scholar] [CrossRef]
- Ruan, Z.; Liu, C.; Guo, Y.; He, Z.; Huang, X.; Jia, X.; Yang, T. SARS-CoV-2 and SARS-CoV: Virtual screening of potential inhibitors targeting RNA-dependent RNA polymerase activity (NSP12). J. Med. Virol. 2021, 93, 389–400. [Google Scholar] [CrossRef]
- Dando, T.M.; Plosker, G.L. Adefovir Dipivoxil. Drugs 2003, 63, 2215–2234. [Google Scholar] [CrossRef]
- Saravolatz, L.D.; Saag, M.S. Emtricitabine, a New Antiretroviral Agent with Activity against HIV and Hepatitis B Virus. Clin. Infect. Dis. 2006, 42, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.-T.; Gish, R.; De Man, R.; Gadano, A.; Sollano, J.; Chao, Y.-C.; Lok, A.; Han, K.-H.; Goodman, Z.; Zhu, J.; et al. A Comparison of Entecavir and Lamivudine for HBeAg-Positive Chronic Hepatitis B. N. Engl. J. Med. 2006, 354, 1001–1010. [Google Scholar] [CrossRef] [Green Version]
- Sheppard, S. Moroxydine: The story of a mislaid antiviral. Acta Derm. -Venereologica. Suppl. 1994, 183, 1–9. [Google Scholar]
- Calleja, C.; Pascussi, J.M.; Mani, J.; Maurel, P.; Vilarem, M. The antibiotic rifampicin is a nonsteroidal ligand and activator of the human glucocorticoid receptor. Nat. Med. 1998, 4, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Gohara, D.; Ha, C.S.; Kumar, S.; Ghosh, B.; Arnold, J.J.; Wisniewski, T.J.; Cameron, C.E. Production of “Authentic” Poliovirus RNA-Dependent RNA Polymerase (3Dpol) by Ubiquitin–Protease-Mediated Cleavage in Escherichia coli. Protein Expr. Purif. 1999, 17, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.A.; Peersen, O.B. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 2004, 23, 3462–3471. [Google Scholar] [CrossRef] [Green Version]
- Hackbart, M.; Deng, X.; Baker, S.C. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc. Natl. Acad. Sci. USA 2020, 117, 8094–8103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Son, K.-N.; Liang, Z.; Lipton, H.L. Double-Stranded RNA Is Detected by Immunofluorescence Analysis in RNA and DNA Virus Infections, Including Those by Negative-Stranded RNA Viruses. J. Virol. 2015, 89, 9383–9392. [Google Scholar] [CrossRef] [Green Version]
- Gunaratne, G.S.; Yang, Y.; Li, F.; Walseth, T.F.; Marchant, J.S. NAADP-dependent Ca2+ signaling regulates Middle East respiratory syndrome-coronavirus pseudovirus translocation through the endolysosomal system. Cell Calcium 2018, 75, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.-L.; Leung, N.; Teo, E.-K.; Tong, M.; Wong, F.; Hann, H.-W.; Han, S.; Poynard, T.; Myers, M.; Chao, G. A 1-Year Trial of Telbivudine, Lamivudine, and the Combination in Patients with Hepatitis B e Antigen—Positive Chronic Hepatitis B. Gastroenterology 2005, 129, 528–536. [Google Scholar] [CrossRef]
- Keeffe, E.B. Liver disease: Hepatitis B. Treatment of chronic hepatitis B With entecavir. Rev. Gastroenterol. Disord. 2006, 6, 112–116. [Google Scholar] [PubMed]
- Bang, L.M.; Scott, L.J. Emtricitabine: An antiretroviral agent for HIV infection. Drugs 2003, 63, 2413–2424, discussion 2425—2416. [Google Scholar] [CrossRef]
- Calvori, C.; Frontali, L.; Leoni, L.; Tecce, G. Effect of Rifamycin on Protein Synthesis. Nat. Cell Biol. 1965, 207, 417–418. [Google Scholar] [CrossRef] [PubMed]





| Name | IC50 (μM) | Max. Dose (μM) | Inhibition % of RdRp Activity at Max. Dose |
|---|---|---|---|
| Remdesivir | 2.585 ± 0.273 | 6.7 | 71.03 |
| Lycorine | 1.465 ± 0.033 | 4.4 | 100 |
| Cepharanthine | >10 | 10 | 5.84 |
| Adefovir Dipivoxil | 3.785 ± 0.866 | 12.5 | 86.98 |
| Emtricitabine | 15.375 ± 3.602 | 100 | 88.21 |
| Telbivudine | 45.928 ± 3.859 | 100 | 76.92 |
| Entecavir Hydrate | 41.993 ± 4.162 | 100 | 83.09 |
| Moroxydine | 48.929 ± 14.370 | 100 | 76.54 |
| Rifampin | 49.434 ± 4.020 | 100 | 96.73 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Min, J.S.; Kwon, S.; Jin, Y.-H. SARS-CoV-2 RdRp Inhibitors Selected from a Cell-Based SARS-CoV-2 RdRp Activity Assay System. Biomedicines 2021, 9, 996. https://doi.org/10.3390/biomedicines9080996
Min JS, Kwon S, Jin Y-H. SARS-CoV-2 RdRp Inhibitors Selected from a Cell-Based SARS-CoV-2 RdRp Activity Assay System. Biomedicines. 2021; 9(8):996. https://doi.org/10.3390/biomedicines9080996
Chicago/Turabian StyleMin, Jung Sun, Sunoh Kwon, and Young-Hee Jin. 2021. "SARS-CoV-2 RdRp Inhibitors Selected from a Cell-Based SARS-CoV-2 RdRp Activity Assay System" Biomedicines 9, no. 8: 996. https://doi.org/10.3390/biomedicines9080996
APA StyleMin, J. S., Kwon, S., & Jin, Y. -H. (2021). SARS-CoV-2 RdRp Inhibitors Selected from a Cell-Based SARS-CoV-2 RdRp Activity Assay System. Biomedicines, 9(8), 996. https://doi.org/10.3390/biomedicines9080996