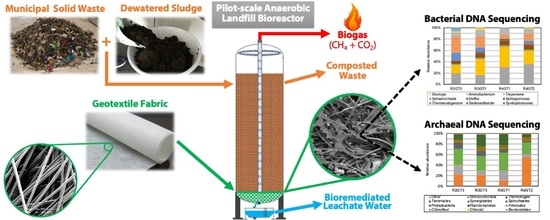
Community Structure of Bacteria and Archaea Associated with Geotextile Filters in Anaerobic Bioreactor Landfills
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bioreactor Setup
2.2. Sample Collection and Scanning Electron Microscopy Analysis
2.3. Metagenomic DNA Extraction
2.4. Bacterial V3–V4 Amplicon Library Preparation and Sequencing
2.5. Archaeal V4 Amplicon Library Preparation and Sequencing
2.6. Processing of Raw Sequences
2.7. Bioinformatics Analysis
2.8. Biodiversity Analysis
2.9. Data Availability
3. Results and Discussion
3.1. SEM Analysis of the Geotextile Fabrics
3.2. Microbial Community Analysis
3.2.1. Bacterial V3–V4 Amplicon Sequencing and Taxonomic Identification
3.2.2. Bacterial Diversity Analysis
3.2.3. Archaeal V4 Amplicon Sequencing and Taxonomic Identification
3.2.4. Archaeal Diversity Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chu, Z.; Wang, W.; Zhou, A.; Huang, W.-C. Charging for municipal solid waste disposal in Beijing. Waste Manag. 2019, 94, 85–94. [Google Scholar] [CrossRef]
- Singh, V.; Phuleria, H.C.; Chandel, M.K. Estimation of energy recovery potential of sewage sludge in India: Waste to watt approach. J. Clean. Prod. 2020, 276, 122538. [Google Scholar] [CrossRef]
- Jayakrishnan, U.; Deka, D.; Das, G. Influence of inoculum variation and nutrient availability on polyhydroxybutyrate production from activated sludge. Int. J. Biol. Macromol. 2020, 163, 2032–2047. [Google Scholar] [CrossRef] [PubMed]
- Broun, R.; Sattler, M. A comparison of greenhouse gas emissions and potential electricity recovery from conventional and bioreactor landfills. J. Clean. Prod. 2016, 112, 2664–2673. [Google Scholar] [CrossRef]
- IEA. Energy Policies of IEA Countries: Turkey; IEA: Paris, France, 2016. [Google Scholar]
- Yaman, C.; Küçükağa, Y.; Pala, B.; Delice, G.; Korkut, N.E.; Akyol, A.; Kara, S. Enhancement of landfill gas production and waste stabilisation by using geotextile filter in a bioreactor landfill. Int. J. Glob. Warm. 2019, 17, 1–22. [Google Scholar] [CrossRef]
- Srivastava, A.N.; Chakma, S. Heavy metals speciation study revealing merits of anaerobic co-disposal of municipal solid waste with discrete paper mill sludges: An experimental investigation in simulated landfill bioreactors. J. Environ. Chem. Eng. 2020, 8, 104337. [Google Scholar] [CrossRef]
- Ali, M.; Yue, D. Population dynamics of microbial species under high and low ammonia nitrogen in the alternate layer bioreactor landfill (ALBL) approach. Bioresour. Technol. 2020, 315, 123787. [Google Scholar] [CrossRef]
- Peces, M.; Astals, S.; Jensen, P.D.; Clarke, W.P. Transition of microbial communities and degradation pathways in anaerobic digestion at decreasing retention time. New Biotechnol. 2021, 60, 52–61. [Google Scholar] [CrossRef]
- Kumar, G.; Reddy, K.R. Rapid stabilization of municipal solid waste in bioreactor landfills: Predictive performance using coupled modeling. Glob. Nest J. 2019, 21, 505–512. [Google Scholar] [CrossRef]
- Rich, C.; Gronow, J.; Voulvoulis, N. The potential for aeration of MSW landfills to accelerate completion. Waste Manag. 2008, 28, 1039–1048. [Google Scholar] [CrossRef]
- Benbelkacem, H.; Bayard, R.; Abdelhay, A.; Zhang, Y.; Gourdon, R. Effect of leachate injection modes on municipal solid waste degradation in anaerobic bioreactor. Bioresour. Technol. 2010, 101, 5206–5212. [Google Scholar] [CrossRef] [PubMed]
- Bilgili, M.S.; Demir, A.; Akkaya, E.; Ozkaya, B. COD fractions of leachate from aerobic and anaerobic pilot scale landfill reactors. J. Hazard. Mater. 2008, 158, 157–163. [Google Scholar] [CrossRef]
- O’Connor, O.A.; Dewan, R.; Galuzzi, P.; Young, L.Y. Landfill leachate: A study of its anaerobic mineralization and toxicity to methanogenesis. Arch. Environ. Contam. Toxicol. 1990, 19, 143–147. [Google Scholar] [CrossRef]
- Pedrazzi, S.; Santunione, G.; Minarelli, A.; Allesina, G. Energy and biochar co-production from municipal green waste gasification: A model applied to a landfill in the north of Italy. Energy Convers. Manag. 2019, 187, 274–282. [Google Scholar] [CrossRef]
- Pohland, F.G.; Kim, J.C. In Situ anaerobic treatment of leachate in landfill bioreactors. Water Sci. Technol. 1999, 40, 203–210. [Google Scholar] [CrossRef]
- Khanal, S.K. Microbiology and biochemistry of anaerobic biotechnology. Anaerob. Biotechnol. Bioenergy Prod. Princ. Appl. 2009, 29–41. [Google Scholar] [CrossRef]
- Gerardi, M.H. The Microbiology of Anaerobic Digesters; John Wiley & Sons: New York, NY, USA, 2003; ISBN 0471468959. [Google Scholar]
- Yaman, C.; Martin, J.P.; Korkut, E. Use of Layered Geotextiles to Provide a Substrate for Biomass Development in Treatment of Septic Tank Effluent Prior to Ground Infiltration. J. Environ. Eng. 2005, 131, 1667–1675. [Google Scholar] [CrossRef]
- Yaman, C.; Küçükağa, Y. Effect of a geotextile filter on leachate quality in a recirculated landfill bioreactor. Desalin. Water Treat. 2017, 80, 61–73. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [Green Version]
- Yaman, C. Improvement of leachate quality and waste stabilization in pilot-scale bioreactor landfills containing geotextile filters and sewage sludge. Desalin. Water Treat. 2019, 158, 41–58. [Google Scholar] [CrossRef] [Green Version]
- Koerner, G.R. Performance Evaluation of Geotextile Filters Used in Leachate Collection Systems of Solid Waste Landfills. Ph.D. Thesis, Drexel University, Philadelphia, PA, USA, 1993. [Google Scholar]
- Coolen, M.J.L.; Muyzer, G.; Rijpstra, W.I.C.; Schouten, S.; Volkman, J.K.; Sinninghe Damsté, J.S. Combined DNA and lipid analyses of sediments reveal changes in Holocene haptophyte and diatom populations in an Antarctic lake. Earth Planet. Sci. Lett. 2004, 223, 225–239. [Google Scholar] [CrossRef]
- Kuczynski, J.; Stombaugh, J.; Walters, W.A.; González, A.; Caporaso, J.G.; Knight, R. Using QIIME to analyze 16s rRNA gene sequences from microbial communities. Curr. Protoc. Microbiol. 2012, 27, 1E–5. [Google Scholar] [CrossRef] [Green Version]
- Comeau, A.M.; Douglas, G.M.; Langille, M.G.I. Microbiome Helper: A Custom and Streamlined Workflow for Microbiome Research. mSystems 2017, 2, e00127-16. [Google Scholar] [CrossRef] [Green Version]
- Cock, P.J.A.; Fields, C.J.; Goto, N.; Heuer, M.L.; Rice, P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2009, 38, 1767–1771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrews, S. FastQC: A quality control tool for high throughput sequence data—ScienceOpen. Babraham Inst. 2010. Available online: https://www.bibsonomy.org/bibtex/f230a919c34360709aa298734d63dca3 (accessed on 5 August 2021).
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, A.; Hannon, G.J. Fastx-Toolkit. FASTQ/A Short-Reads Preprocessing Tools (Unpublished). Available online: http://hannonlab.cshl.edu/fastx_toolkit (accessed on 10 October 2020).
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 2016, e2584. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pẽa, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [Green Version]
- Chao, A. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar]
- Lozupone, C.; Lladser, M.E.; Knights, D.; Stombaugh, J.; Knight, R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011, 5, 169–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azari, M.; Walter, U.; Rekers, V.; Gu, J.D.; Denecke, M. More than a decade of experience of landfill leachate treatment with a full-scale anammox plant combining activated sludge and activated carbon biofilm. Chemosphere 2017, 174, 117–126. [Google Scholar] [CrossRef]
- Stamps, B.W.; Lyles, C.N.; Suflita, J.M.; Masoner, J.R.; Cozzarelli, I.M.; Kolpin, D.W.; Stevenson, B.S. Municipal solid waste landfills harbor distinct microbiomes. Front. Microbiol. 2016, 7, 534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sang, N.N.; Soda, S.; Sei, K.; Ike, M. Effect of Aeration on Stabilization of Organic Solid Waste and Microbial Population Dynamics in Lab-Scale Landfill Bioreactors. J. Biosci. Bioeng. 2008, 106, 425–432. [Google Scholar] [CrossRef]
- Thauer, R.K.; Kaster, A.K.; Seedorf, H.; Buckel, W.; Hedderich, R. Methanogenic archaea: Ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 2008, 6, 579–591. [Google Scholar] [CrossRef]
- Kougias, P.G.; Angelidaki, I. Biogas and its opportunities—A review. Front. Environ. Sci. Eng. 2018, 12, 14. [Google Scholar] [CrossRef]
- Köchling, T.; Sanz, J.L.; Gavazza, S.; Florencio, L. Analysis of microbial community structure and composition in leachates from a young landfill by 454 pyrosequencing. Appl. Microbiol. Biotechnol. 2015, 99, 5657–5668. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Wang, Y.; Tang, W.; Lei, Y. Bacterial community diversity in municipal waste landfill sites. Appl. Microbiol. Biotechnol. 2015, 99, 7745–7756. [Google Scholar] [CrossRef]
- Laloui-Carpentier, W.; Li, T.; Vigneron, V.; Mazéas, L.; Bouchez, T. Methanogenic diversity and activity in municipal solid waste landfill leachates. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2006, 89, 423–434. [Google Scholar] [CrossRef]
- Ziegler, A.S.; McIlroy, S.J.; Larsen, P.; Albertsen, M.; Hansen, A.A.; Heinen, N.; Nielsen, P.H. Dynamics of the fouling layer microbial community in a membrane bioreactor. PLoS ONE 2016, 11, e0158811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Söllinger, A.; Urich, T. Methylotrophic methanogens everywhere—Physiology and ecology of novel players in global methane cycling. Biochem. Soc. Trans. 2019, 47, 1895–1907. [Google Scholar] [CrossRef] [PubMed]
- Zinder, S.H. Physiological Ecology of Methanogens. In Methanogenesis; Springer: Cham, Switzerland, 1993; pp. 128–206. [Google Scholar]
- Qu, X.; Mazéas, L.; Vavilin, V.A.; Epissard, J.; Lemunier, M.; Mouchel, J.M.; He, P.J.; Bouchez, T. Combined monitoring of changes in δ13CH4 and archaeal community structure during mesophilic methanization of municipal solid waste. FEMS Microbiol. Ecol. 2009, 68, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Jayanthi, B.; Emenike, C.U.; Agamuthu, P.; Simarani, K.; Mohamad, S.; Fauziah, S.H. Selected microbial diversity of contaminated landfill soil of Peninsular Malaysia and the behavior towards heavy metal exposure. Catena 2016, 147, 25–31. [Google Scholar] [CrossRef]
- Karelová, E.; Harichová, J.; Stojnev, T.; Pangallo, D.; Ferianc, P. The isolation of heavy-metal resistant culturable bacteria and resistance determinants from a heavy-metal-contaminated site. Biologia 2011, 66, 18–26. [Google Scholar] [CrossRef]
- Zainun, M.Y.; Simarani, K. Metagenomics profiling for assessing microbial diversity in both active and closed landfills. Sci. Total Environ. 2018, 616–617, 269–278. [Google Scholar] [CrossRef]








| Name | Type and Model | Thickness (mm) | Apparent Opening Size | Permeability (L m−2 s−1) | Production Material | Production Method |
|---|---|---|---|---|---|---|
| R4GT1 | TenCate TS50 | 1.91 | 112 | 113 | Polypropylene | Needle-punched |
| R4GT2 | GeoTeknik 200 | 1.40 | 84 | 60 | (PP) | Needle-punched |
| R3GT3 and R2GT3 | IzoTeknik 200 | 1.90 | 110 | 95 | Polyester | Needle-punched |
| Sample ID | Reactor | Position in the Drainage Chamber | Reactor Feed |
|---|---|---|---|
| R2GT3 | R2 | Middle | Solid Waste + Sewage Sludge |
| R3GT3 | R3 | Middle | Solid Waste |
| R4GT2 | R4 | Upper | Solid Waste |
| R4GT1 | R4 | Lower | Solid Waste |
| Sample | Raw PE | Combined | Qualified | Nochime | AvgLen (nt) | Q20 (%) | Q30 (%) | GC Content (%) | Effective Rate (%) |
|---|---|---|---|---|---|---|---|---|---|
| R2GT3 | 99,376 | 90,283 | 80,243 | 77,978 | 415 | 98.19 | 96.23 | 53.78 | 78.47 |
| R3GT3 | 84,324 | 76,769 | 68,397 | 65,422 | 413 | 98.20 | 96.27 | 54.38 | 77.58 |
| R4GT2 | 98,288 | 89,942 | 80,499 | 77,708 | 415 | 98.23 | 96.29 | 54.30 | 79.06 |
| R4GT1 | 98,918 | 91,918 | 82,881 | 80,885 | 409 | 98.26 | 96.33 | 57.30 | 81.77 |
| Sample | Raw PE | Combined | Qualified | Nochime | AvgLen (nt) | Q20 (%) | Q30 (%) | GC Content (%) | Effective Rate (%) |
|---|---|---|---|---|---|---|---|---|---|
| R2GT3 | 131,103 | 113,360 | 99,611 | 96,773 | 380 | 97.30 | 94.50 | 53.65 | 73.81 |
| R3GT3 | 101,791 | 87,520 | 77,144 | 74,432 | 381 | 97.33 | 94.55 | 54.50 | 73.12 |
| R4GT2 | 58,195 | 47,640 | 40,499 | 39,024 | 380 | 96.84 | 93.57 | 53.92 | 67.06 |
| R4GT1 | 45,258 | 34,062 | 28,378 | 28,378 | 383 | 96.57 | 96.57 | 55.75 | 58.96 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yaman, C.; Rehman, S.; Ahmad, T.; Kucukaga, Y.; Pala, B.; AlRushaid, N.; Riyaz Ul Hassan, S.; Yaman, A.B. Community Structure of Bacteria and Archaea Associated with Geotextile Filters in Anaerobic Bioreactor Landfills. Processes 2021, 9, 1377. https://doi.org/10.3390/pr9081377
Yaman C, Rehman S, Ahmad T, Kucukaga Y, Pala B, AlRushaid N, Riyaz Ul Hassan S, Yaman AB. Community Structure of Bacteria and Archaea Associated with Geotextile Filters in Anaerobic Bioreactor Landfills. Processes. 2021; 9(8):1377. https://doi.org/10.3390/pr9081377
Chicago/Turabian StyleYaman, Cevat, Suriya Rehman, Tanveer Ahmad, Yusuf Kucukaga, Burcu Pala, Noor AlRushaid, Syed Riyaz Ul Hassan, and Ayse Burcu Yaman. 2021. "Community Structure of Bacteria and Archaea Associated with Geotextile Filters in Anaerobic Bioreactor Landfills" Processes 9, no. 8: 1377. https://doi.org/10.3390/pr9081377
APA StyleYaman, C., Rehman, S., Ahmad, T., Kucukaga, Y., Pala, B., AlRushaid, N., Riyaz Ul Hassan, S., & Yaman, A. B. (2021). Community Structure of Bacteria and Archaea Associated with Geotextile Filters in Anaerobic Bioreactor Landfills. Processes, 9(8), 1377. https://doi.org/10.3390/pr9081377