Effect of Functional Groups on Protein Adsorption Performance of Membrane Adsorbers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
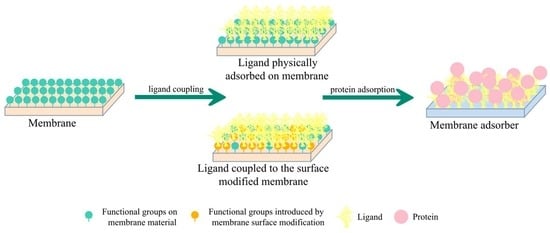
2.2. Preparation and Characterization of MA
2.2.1. Ligand Physically Adsorbed on Membrane
2.2.2. Alkali Treatment
2.2.3. Oxidation Treatment
2.2.4. PDA deposition Modification
2.3. Characterization Methods
2.4. MA Performance Testing
3. Results and Discussion
3.1. Protein Adsorption/Desorption Performance on Different Membrane Functional Groups
3.1.1. Effect of Membrane Functional Groups
3.1.2. Effect of New Functional Groups Introduced by Alkali/Oxidation Treatment
3.1.3. Effect of New Functional Groups Introduced by PDA Deposition Treatment
3.2. Optimization of the Preparation Conditions of the PEI@PVDF-PDA
3.2.1. Effect of the pH Value of the Dopamine Polymerization Process
3.2.2. Effect of the Temperature of Dopamine Polymerization Process
3.2.3. Effect of Ligand Molecular Weight
3.2.4. Effect of Ligand Concentration
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ghosh, R. Protein separation using membrane chromatography: Opportunities and challenges. J. Chromatogr. A 2002, 952, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Abdulhussain, N.; Nawada, S.; Schoenmakers, P. Latest Trends on the Future of Three-Dimensional Separations in Chromatography. Chem. Rev. 2021, 121, 12016–12034. [Google Scholar] [CrossRef] [PubMed]
- Brandt, S.; Goffe, R.A.; Kessler, S.B.; O’Connor, J.L.; Zale, S.E. Membrane-Based Affinity Technology for Commercial Scale Purifications. Nat. Biotechnol. 1988, 6, 779–782. [Google Scholar] [CrossRef]
- Orr, V.; Zhong, L.; Moo-Young, M.; Chou, C.P. Recent advances in bioprocessing application of membrane chromatography. Biotechnol. Adv. 2013, 31, 450–465. [Google Scholar] [CrossRef]
- Khan, M.K.; Luo, J.; Khan, R.; Fan, J.; Wan, Y. Facile and green fabrication of cation exchange membrane adsorber with unprecedented adsorption capacity for protein purification. J. Chromatogr. A 2017, 1521, 19–26. [Google Scholar] [CrossRef]
- Wang, C.; Shen, J.; Zhu, J.; Bo, C.; Wei, Y. Tetrazole-functionalized cation-exchange membrane adsorbers with high binding capacity and unique separation feature for protein. J. Chromatogr. B 2018, 1097–1098, 18–26. [Google Scholar] [CrossRef]
- Chenette, H.C.; Robinson, J.R.; Hobley, E.; Husson, S.M. Development of high-productivity, strong cation-exchange adsorbers for protein capture by graft polymerization from membranes with different pore sizes. J. Membr. Sci. 2012, 423–424, 43–52. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Gurgel, P.V.; Carbonell, R.G. Preparation and characterization of anion exchange adsorptive nonwoven membranes with high protein binding capacity. J. Membr. Sci. 2015, 493, 349–359. [Google Scholar] [CrossRef]
- Ma, N.; Yao, D.; Yang, H.; Yin, J.; Wang, H.; Zhang, Y.; Meng, J. Surface Modification of Cellulose Membranes To Prepare a High-Capacity Membrane Adsorber for Monoclonal Antibody Purification via Hydrophobic Charge-Induction Chromatography. Ind. Eng. Chem. Res. 2018, 57, 13235–13246. [Google Scholar] [CrossRef]
- Ho, C.-C.; Prodan, B.N.; Kiessling, B.; Co, C.C. Effects of membrane surface chemistry on fouling investigated using self-assembled monolayers. J. Membr. Sci. 2011, 366, 342–348. [Google Scholar] [CrossRef]
- Saeki, D.; Yonamine, G.; Matsuyama, H. Effect of hydrophilic polymer modification of reverse osmosis membrane surfaces on organic adsorption and biofouling behavior. Colloids Surf. A 2020, 609, 125680. [Google Scholar] [CrossRef]
- Bagheri, M.; Mirbagheri, S.A. Critical review of fouling mitigation strategies in membrane bioreactors treating water and wastewater. Bioresour. Technol. 2018, 258, 318–334. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liao, B.-Q.; Zhou, X.; He, Y.; Hong, H.; Lin, H.; Chen, J. Effects of hydrophilicity/hydrophobicity of membrane on membrane fouling in a submerged membrane bioreactor. Bioresour. Technol. 2015, 175, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yu, S.; Gao, C.; Feng, X. Surface modification of thin film composite polyamide membranes by electrostatic self deposition of polycations for improved fouling resistance. Sep. Purif. Technol. 2009, 66, 287–294. [Google Scholar] [CrossRef]
- Wang, Q.; Wei, X.; Wang, G.-R.; Lu, T.-D.; Shi, Q.; Sun, S.-P. Inner-selective coordination nanofiltration hollow fiber membranes from assist-pressure modified substrate. J. Membr. Sci. 2021, 626, 119186. [Google Scholar] [CrossRef]
- Sagitha, P.; Reshmi, C.; Sundaran, S.P.; Sujith, A. Recent advances in post-modification strategies of polymeric electrospun membranes. Eur. Polym. J. 2018, 105, 227–249. [Google Scholar] [CrossRef]
- Ye, J.; Chu, J.; Yin, J.; Zhang, Y.; Meng, J. Surface modification of regenerated cellulose membrane based on thiolactone chemistry—A novel platform for mixed mode membrane adsorbers. Appl. Surf. Sci. 2020, 511, 145539. [Google Scholar] [CrossRef]
- He, D.; Ulbricht, M. Preparation and characterization of porous anion-exchange membrane adsorbers with high protein-binding capacity. J. Membr. Sci. 2008, 315, 155–163. [Google Scholar] [CrossRef]
- Fan, J.; Luo, J.; Chen, X.; Wan, Y. Facile preparation of salt-tolerant anion-exchange membrane adsorber using hydrophobic membrane as substrate. J. Chromatogr. A 2017, 1490, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Luo, J.; Song, W.; Wan, Y. One-step purification of α1-antitrypsin by regulating polyelectrolyte ligands on mussel-inspired membrane adsorber. J. Membr. Sci. 2017, 528, 155–162. [Google Scholar] [CrossRef]
- Shi, H.; Xue, L.; Gao, A.; Fu, Y.; Zhou, Q.; Zhu, L. Fouling-resistant and adhesion-resistant surface modification of dual layer PVDF hollow fiber membrane by dopamine and quaternary polyethyleneimine. J. Membr. Sci. 2016, 498, 39–47. [Google Scholar] [CrossRef]
- Choi, K.I.; Lee, J.H. Preparation and dispersion of metal oxide nanostructures using amino acid-assisted chemical routes: An overview. Sci. Adv. Mater. 2011, 3, 811–820. [Google Scholar] [CrossRef]
- Carlton, L.; Staskun, B.; van Es, T. A nitrogen-15 NMR study of hydrogen bonding in 1-alkyl-4-imino-1,4-dihydro-3-quinolinecarboxylic acids and related compounds. Magn. Reson. Chem. 2006, 44, 510–514. [Google Scholar] [CrossRef] [PubMed]
- Kolev, S.K.; St Petkov, P.; Rangelov, M.A.; Vayssilov, G.N. Density functional study of hydrogen bond formation between methanol and organic molecules containing Cl, F, NH2, OH, and COOH functional groups. J. Phys. Chem. A 2011, 115, 14054–14068. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.-M.; Chen, C.-I.; Kao, P.-M.; Chen, H.-B.; Huang, H.-C.; Yao, C.-J.; Liu, Y.-C. Preparation of the immobilized metal affinity membrane with high amount of metal ions and protein adsorption efficiencies. Process. Biochem. 2010, 45, 500–506. [Google Scholar] [CrossRef]
- Ramamoorthy, S.K.; Bakare, F.; Herrmann, R.; Skrifvars, M. Performance of biocomposites from surface modified regenerated cellulose fibers and lactic acid thermoset bioresin. Cellulose 2015, 22, 2507–2528. [Google Scholar] [CrossRef] [Green Version]
- Ross, G.; Watts, J.; Hill, M.; Morrissey, P. Surface modification of poly(vinylidene fluoride) by alkaline treatment1. The degradation mechanism. Polymer 2000, 41, 1685–1696. [Google Scholar] [CrossRef]
- Kumar, R.; Ismail, A.; Kassim, M.; Isloor, A.M. Modification of PSf/PIAM membrane for improved desalination applications using Chitosan coagulation media. Desalination 2013, 317, 108–115. [Google Scholar] [CrossRef]
- Kim, K.; Lee, K.; Cho, K.; Park, C. Surface modification of polysulfone ultrafiltration membrane by oxygen plasma treatment. J. Membr. Sci. 2002, 199, 135–145. [Google Scholar] [CrossRef]
- Liang, S.; Kang, Y.; Tiraferri, A.; Giannelis, E.P.; Huang, X.; Elimelech, M. Highly hydrophilic polyvinylidene fluoride (PVDF) ultrafiltration membranes via postfabrication grafting of surface-tailored silica nanoparticles. ACS. Appl. Mater. Interfaces 2013, 5, 6694–6703. [Google Scholar] [CrossRef]
- Xiong, Z.; Zhong, Y.; Lin, H.; Liu, F.; Li, T.; Li, J. PDLA/PLLA ultrafiltration membrane with excellent permeability, rejection and fouling resistance via stereocomplexation. J. Membr. Sci. 2017, 533, 103–111. [Google Scholar] [CrossRef]
- Liu, Y.; Ai, K.; Lu, L. Polydopamine and Its Derivative Materials: Synthesis and Promising Applications in Energy, Environmental, and Biomedical Fields. Chem. Rev. 2014, 114, 5057–5115. [Google Scholar] [CrossRef] [PubMed]
- Ball, V.; Del Frari, D.; Toniazzo, V.; Ruch, D. Kinetics of polydopamine film deposition as a function of pH and dopamine concentration: Insights in the polydopamine deposition mechanism. J. Colloid Interface Sci. 2012, 386, 366–372. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Zhu, L.; Zhu, L.; Zhu, B.; Xu, Y. Surface Characteristics of a Self-Polymerized Dopamine Coating Deposited on Hydrophobic Polymer Films. Langmuir 2011, 27, 14180–14187. [Google Scholar] [CrossRef]
- Yu, L.-L.; Tao, S.-P.; Dong, X.-Y.; Sun, Y. Protein adsorption to poly(ethylenimine)-modified Sepharose FF: I. A critical ionic capacity for drastically enhanced capacity and uptake kinetics. J. Chromatogr. A 2013, 1305, 76–84. [Google Scholar] [CrossRef] [PubMed]









Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Yang, J.; Yan, Y.; Zhang, W. Effect of Functional Groups on Protein Adsorption Performance of Membrane Adsorbers. Separations 2023, 10, 211. https://doi.org/10.3390/separations10030211
Zhang L, Yang J, Yan Y, Zhang W. Effect of Functional Groups on Protein Adsorption Performance of Membrane Adsorbers. Separations. 2023; 10(3):211. https://doi.org/10.3390/separations10030211
Chicago/Turabian StyleZhang, Lifang, Jialin Yang, Yiqing Yan, and Weidong Zhang. 2023. "Effect of Functional Groups on Protein Adsorption Performance of Membrane Adsorbers" Separations 10, no. 3: 211. https://doi.org/10.3390/separations10030211
APA StyleZhang, L., Yang, J., Yan, Y., & Zhang, W. (2023). Effect of Functional Groups on Protein Adsorption Performance of Membrane Adsorbers. Separations, 10(3), 211. https://doi.org/10.3390/separations10030211