A Rapid and Sensitive UPLC-MS/MS Method for Quantifying Capmatinib in Human Liver Microsomes: Evaluation of Metabolic Stability by In Silico and In Vitro Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Instruments
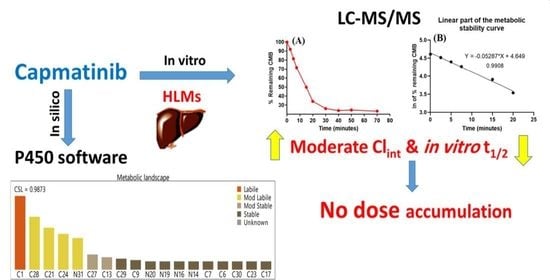
2.3. In Silico Analysis of the Metabolic Lability of CMB
2.4. Optimization of UPLC-MS/MS Parameters
2.5. Preparation of Working Dilutions of CMB and PMT
2.6. Construction of CMB Calibration Curve
2.7. Extraction of CMB and PMT from the HLM Matrix
2.8. Validation of the UPLC-MS/MS Method
2.8.1. Specificity
2.8.2. Sensitivity and Linearity
2.8.3. Accuracy and Precision
2.8.4. Extraction Recovery and Matrix Effect
2.8.5. Stability
2.9. In Vitro Evaluation of CMB Metabolic Stability
3. Results and Discussions
3.1. In Silico Analysis of the Metabolic Stability of CMB
3.2. Establishment of the UPLC-MS/MS Analytical Method
3.3. Validation of the Established LC-MS/MS Method
3.3.1. Specificity
3.3.2. Sensitivity and Linearity
3.3.3. Precision and Accuracy
3.3.4. HLM Matrix Does Not Influence the Extraction or Recovery of CMB with the UPLC-MS/MS Chromatographic Method
3.3.5. CMB was Stable in the Stock Solution and HLMs Matrix
3.4. In Vitro Metabolic Stability of CMB
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mattiuzzi, C.; Lippi, G. Current Cancer Epidemiology. J. Epidemiol. Glob. Health 2019, 9, 217–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jemal, A.; Siegel, R.; Ward, E.; Hao, Y.; Xu, J.; Murray, T.; Thun, M.J. Cancer statistics, 2008. CA A Cancer J. Clin. 2008, 58, 71–96. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Siegel, R.L.; Jemal, A. Lung cancer statistics. In Lung Cancer and Personalized Medicine; Springer: Berlin/Heidelberg, Germany, 2016; pp. 1–19. [Google Scholar]
- Chmielecki, J.; Foo, J.; Oxnard, G.R.; Hutchinson, K.; Ohashi, K.; Somwar, R.; Wang, L.; Amato, K.R.; Arcila, M.; Sos, M.L. Optimization of dosing for EGFR-mutant non–small cell lung cancer with evolutionary cancer modeling. Sci. Transl. Med. 2011, 3, 90ra59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katayama, R.; Khan, T.M.; Benes, C.; Lifshits, E.; Ebi, H.; Rivera, V.M.; Shakespeare, W.C.; Iafrate, A.J.; Engelman, J.A.; Shaw, A.T. Therapeutic strategies to overcome crizotinib resistance in non-small cell lung cancers harboring the fusion oncogene EML4-ALK. Proc. Natl. Acad. Sci. USA 2011, 108, 7535–7540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barinaga, M. From Bench Top to Bedside. Science 1997, 278, 1036–1039. [Google Scholar] [CrossRef] [PubMed]
- Shenouda, S.K.; Alahari, S.K. MicroRNA function in cancer: Oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009, 28, 369. [Google Scholar] [CrossRef]
- Sechler, M.; Cizmic, A.D.; Avasarala, S.; Van Scoyk, M.; Brzezinski, C.; Kelley, N.; Bikkavilli, R.K.; Winn, R.A. Non-small-cell lung cancer: Molecular targeted therapy and personalized medicine–drug resistance, mechanisms, and strategies. Pharm. Pers. Med. 2013, 6, 25. [Google Scholar]
- Cheng, L.; Alexander, R.E.; MacLennan, G.T.; Cummings, O.W.; Montironi, R.; Lopez-Beltran, A.; Cramer, H.M.; Davidson, D.D.; Zhang, S. Molecular pathology of lung cancer: Key to personalized medicine. Mod. Pathol. 2012, 25, 347–369. [Google Scholar] [CrossRef] [Green Version]
- Reungwetwattana, T.; Liang, Y.; Zhu, V.; Ou, S.-H.I. The race to target MET exon 14 skipping alterations in non-small cell lung cancer: The why, the how, the who, the unknown, and the inevitable. Lung Cancer 2017, 103, 27–37. [Google Scholar] [CrossRef] [Green Version]
- Kunii, K.; Davis, L.; Gorenstein, J.; Hatch, H.; Yashiro, M.; Di Bacco, A.; Elbi, C.; Lutterbach, B. FGFR2-amplified gastric cancer cell lines require FGFR2 and Erbb3 signaling for growth and survival. Cancer Res. 2008, 68, 2340–2348. [Google Scholar] [CrossRef] [Green Version]
- Garon, E.B.; Brodrick, P. Targeted Therapy Approaches for MET Abnormalities in Non-Small Cell Lung Cancer. Drugs 2021, 81, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, S. Capmatinib: First approval. Drugs 2020, 80, 1125–1131. [Google Scholar] [CrossRef] [PubMed]
- Vansteenkiste, J.F.; Van De Kerkhove, C.; Wauters, E.; Van Mol, P. Capmatinib for the treatment of non-small cell lung cancer. Expert Rev. Anticancer Ther. 2019, 19, 659–671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glaenzel, U.; Jin, Y.; Hansen, R.; Schroer, K.; Rahmanzadeh, G.; Pfaar, U.; van Lier, J.J.; Borell, H.; Meissner, A.; Camenisch, G. Absorption, distribution, metabolism, and excretion of capmatinib (INC280) in healthy male volunteers and in vitro aldehyde oxidase phenotyping of the major metabolite. Drug Metab. Dispos. 2020, 48, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Cui, X.; Pognan, N.; Quinlan, M.; Kapoor, S.; Rahmanzadeh, G.; Giovannini, M.; Marbury, T.C. Pharmacokinetics of capmatinib in participants with hepatic impairment: A phase 1, open-label, single-dose, parallel-group study. Br. J. Clin. Pharmacol. 2022, 88, 91–102. [Google Scholar] [CrossRef]
- Fan, X.; Yang, G.; Cui, W.; Liu, Q.; Zhang, Z.; Zhang, Z. Development and full validation of an LC–MS/MS methodology to quantify capmatinib (INC280) following intragastric administration to rats. Biomed. Chromatogr. 2020, 34, e4768. [Google Scholar] [CrossRef]
- Zhou, C.; Tian, J.; Lin, P.; Liu, T.; He, A.; Fang, L.; Sun, L. Quantitation of capmatinib, a mesenchymal-epithelial transition factor inhibitor by UPLC-MS/MS in rat plasma and its application to a pharmacokinetic study. Bioanalysis 2020, 12, 285–293. [Google Scholar] [CrossRef]
- Zayed, A.; Jaber, S.A.A.; Al Hroot, J.; Hawamdeh, S.; Ayoub, N.M.; Qinna, N.A. HPLC with Fluorescence and Photodiode Array Detection for Quantifying Capmatinib in Biological Samples: Application to In Vivo and In Vitro Studies. Molecules 2022, 27, 8582. [Google Scholar] [CrossRef]
- Tyzack, J.D.; Kirchmair, J. Computational methods and tools to predict cytochrome P450 metabolism for drug discovery. Chem. Biol. Drug Des. 2019, 93, 377–386. [Google Scholar] [CrossRef]
- Marothu Vamsi, K.; Kantamaneni, P.; Gorrepati, M. In vitro Metabolic Stability of Drugs and Applications of LC-MS in Metabolite Profiling. In Drug Metabolism; Katherine, D., Ed.; IntechOpen: Rijeka, Croatia, 2021; p. 77. [Google Scholar]
- Houston, J.B. Utility of in vitro drug metabolism data in predicting in vivo metabolic clearance. Biochem. Pharm. 1994, 47, 1469–1479. [Google Scholar] [CrossRef]
- Obach, R.S.; Baxter, J.G.; Liston, T.E.; Silber, B.M.; Jones, B.C.; MacIntyre, F.; Rance, D.J.; Wastall, P. The prediction of human pharmacokinetic parameters from preclinical and in vitro metabolism data. J. Pharm. Exp. 1997, 283, 46–58. [Google Scholar]
- Attwa, M.W.; Kadi, A.A.; Darwish, H.W.; Amer, S.M.; Alrabiah, H. A reliable and stable method for the determination of foretinib in human plasma by LC-MS/MS: Application to metabolic stability investigation and excretion rate. Eur. J. Mass Spectrom. 2018, 24, 344–351. [Google Scholar] [CrossRef] [PubMed]
- Darwish, H.W.; Kadi, A.A.; Attwa, M.W.; Almutairi, H.S. Investigation of metabolic stability of the novel ALK inhibitor brigatinib by liquid chromatography tandem mass spectrometry. Clin. Chim. Acta 2018, 480, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Attwa, M.W.; Darwish, H.W.; Alhazmi, H.A.; Kadi, A.A. Investigation of metabolic degradation of new ALK inhibitor: Entrectinib by LC-MS/MS. Clin. Chim. Acta 2018, 485, 298–304. [Google Scholar] [CrossRef]
- Amer, S.M.; Kadi, A.A.; Darwish, H.W.; Attwa, M.W. LC–MS/MS method for the quantification of masitinib in RLMs matrix and rat urine: Application to metabolic stability and excretion rate. Chem. Cent. J. 2017, 11, 136. [Google Scholar] [CrossRef] [Green Version]
- Alrabiah, H.; Kadi, A.A.; Attwa, M.W.; Abdelhameed, A.S. A simple liquid chromatography-tandem mass spectrometry method to accurately determine the novel third-generation EGFR-TKI naquotinib with its applicability to metabolic stability assessment. RSC Adv. 2019, 9, 4862–4869. [Google Scholar] [CrossRef] [Green Version]
- Kadi, A.A.; Darwish, H.W.; Abuelizz, H.A.; Alsubi, T.A.; Attwa, M.W. Identification of reactive intermediate formation and bioactivation pathways in Abemaciclib metabolism by LC-MS/MS: In vitro metabolic investigation. R. Soc. Open Sci. 2019, 6, 181714. [Google Scholar] [CrossRef] [Green Version]
- Attwa, M.W.; Kadi, A.A.; Abdelhameed, A.S.; Alhazmi, H.A. Metabolic stability assessment of new parp inhibitor talazoparib using validated lc–ms/ms methodology: In silico metabolic vulnerability and toxicity studies. Drug Des. Dev. Ther. 2020, 14, 783–793. [Google Scholar] [CrossRef]
- Busby, W.F.; Ackermann, J.M.; Crespi, C.L. Effect of methanol, ethanol, dimethyl sulfoxide, and acetonitrile on in vitro activities of cDNA-expressed human cytochromes P-450. Drug Metab. Dispos. 1999, 27, 246–249. [Google Scholar]
- Störmer, E.; Roots, I.; Brockmöller, J. Benzydamine N-oxidation as an index reaction reflecting FMO activity in human liver microsomes and impact of FMO3 polymorphisms on enzyme activity. Br. J. Clin. Pharmacol. 2000, 50, 553–561. [Google Scholar] [CrossRef] [Green Version]
- Fouin-Fortunet, H.; Tinel, M.; Descatoire, V.; Letteron, P.; Larrey, D.; Geneve, J.; Pessayre, D. Inactivation of cytochrome P-450 by the drug methoxsalen. J. Pharmacol. Exp. Ther. 1986, 236, 237–247. [Google Scholar] [PubMed]
- United State of America–Food and Drug Administration. Bioanalytical Method Validation Guidance for Industry. 2018. Available online: https://www.fda.gov/ucm/groups/fdagov-public/@fdagov-drugsgen/documents/document/ucm070107.pdf (accessed on 2 February 2022).
- Smith, G. European Medicines Agency guideline on bioanalytical method validation: What more is there to say? Bioanalysis 2012, 4, 865–868. [Google Scholar] [CrossRef] [PubMed]
- McNaney, C.A.; Drexler, D.M.; Hnatyshyn, S.Y.; Zvyaga, T.A.; Knipe, J.O.; Belcastro, J.V.; Sanders, M. An automated liquid chromatography-mass spectrometry process to determine metabolic stability half-life and intrinsic clearance of drug candidates by substrate depletion. Assay Drug Dev. Technol. 2008, 6, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Słoczyńska, K.; Gunia-Krzyżak, A.; Koczurkiewicz, P.; Wójcik-Pszczoła, K.; Żelaszczyk, D.; Popiół, J.; Pękala, E. Metabolic stability and its role in the discovery of new chemical entities. Acta Pharm. 2019, 69, 345–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirchmair, J.; Williamson, M.J.; Tyzack, J.D.; Tan, L.; Bond, P.J.; Bender, A.; Glen, R.C. Computational prediction of metabolism: Sites, products, SAR, P450 enzyme dynamics, and mechanisms. J. Chem. Inf. Model 2012, 52, 617–648. [Google Scholar] [CrossRef] [PubMed]
- Hunt, P.A.; Segall, M.D.; Tyzack, J.D. WhichP450: A multi-class categorical model to predict the major metabolising CYP450 isoform for a compound. J. Comput.—Aided Mol. Des. 2018, 32, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.G.; Le, H.; Khojasteh, C.; ECA Hop, C. Comparison of metabolic soft spot predictions of CYP3A4, CYP2C9 and CYP2D6 substrates using MetaSite and StarDrop. Comb. Chem. High Throughput Screen. 2011, 14, 811–823. [Google Scholar] [CrossRef]
- Leahy, D.E. Integrating invitro ADMET data through generic physiologically based pharmacokinetic models. Expert Opin. Drug Metab. Toxicol. 2006, 2, 619–628. [Google Scholar] [CrossRef]





| Acquity UPLC (H10UPH) | Acquity TQD MS (QBB1203) | ||
|---|---|---|---|
| Isocratic mobile phase | 45% ACN | ESI | Positive ESI |
| 0.1% HCOOH in H2O (55%; pH: 3.2) | Nitrogen (drying gas; 350 °C) at 100 L/H flow rate | ||
| Flow rate: 0.15 mL/min | Cone gas: 100 L/H flow rate | ||
| Injection volume: 5.0 μL | Voltage of extractor: 3.0 (V) | ||
| ZORBAX Eclipse plus-C18 column | 50.0 mm long | Voltage of RF lens: 0.1 (V) | |
| 2.1 mm i.d. | Capillary voltage: 4 KV | ||
| 1.8 μm particle size | Collision cell | Argon gas (collision gas) at 0.14 mL/min flow rate | |
| T: 22.0 ± 2.0 °C | Mode | MRM | |
| Time | Retention Time | MRM Transitions | ||
|---|---|---|---|---|
| Mass spectra segment | 0.0 to 1.5 min | CMB (0.97 min) | Qualification traces (m/z) | 413→354 CE a: 6 and CV b: 26 |
| Quantification traces (m/z) | 413→ 82 CE: 6 and CV: 32 | |||
| 1.5 to 3.0 min | PMT (IS; 1.98 min) | Quantification traces (m/z) | 488→401 CE: 26 and CV: 36 | |
| Qualification traces (m/z) | 488→186 CE: 16 and CV: 26 | |||
| Analytes | Mobile Phase | Extraction Method | Stationary Phase | |||
|---|---|---|---|---|---|---|
| ACN (45%) | Methanol | Protein precipitation using ACN | Solid phase extraction | C18 column | C8 column | |
| CMB | 0.97 min | 1.35 min | High recovery (99.28%) | Low recovery (75.87%) | 0.97 min | 1.68 min |
| Good peak shape | Tailed peaks | Precise results (RSD < 4.30%) | Not precise | Perfect shape | Tailed peaks | |
| PMT | 1.97 min | 1.57 min | High recovery (101.7%) | Low recovery (82.49 %) | 1.97 min | 1.24 min |
| Good peak shape | Overlapped | Precise results (RSD < 4.40%) | Not precise | Perfect shape | Perfect shape | |
| CMB (ng/mL) | Mean | SD | RSD (%) | Accuracy (%) | Recovery |
|---|---|---|---|---|---|
| 1.0 | 0.94 | 0.01 | 1.29 | −6.39 | 93.61 |
| 15.0 | 15.14 | 0.18 | 1.17 | 0.92 | 100.92 |
| 50.0 | 50.93 | 1.99 | 3.92 | 1.85 | 101.85 |
| 200.0 | 199.16 | 3.45 | 1.73 | −0.42 | 99.58 |
| 400.0 | 405.84 | 1.98 | 0.49 | 1.46 | 101.46 |
| 500.0 | 503.70 | 5.13 | 1.02 | 0.74 | 100.74 |
| 1500.0 | 1520.02 | 18.31 | 1.20 | 1.33 | 101.33 |
| 3000.0 | 2958.63 | 26.24 | 0.89 | −1.38 | 98.62 |
| % Recovery | 99.76 ± 2.71 |
| CMB (ng/mL) | Intra-Day Assay (Twelve Repeats on the Same Day) | Inter-Day Assay (Six Repeats on Three Following Days) | ||||||
|---|---|---|---|---|---|---|---|---|
| QCs | 1.0 (LLOQ) | 3.0 (LQC) | 900.0 (MQC) | 2400.0 (HQC) | 1.0 (LLOQ) | 3.0 (LQC) | 900.0 (MQC) | 2400.0 (HQC) |
| Average | 0.94 | 3.07 | 911.07 | 2407.67 | 0.92 | 3.13 | 915.48 | 2359.96 |
| SD | 0.01 | 0.11 | 4.18 | 22.39 | 0.02 | 0.22 | 5.15 | 22.37 |
| Precision (%RSD) | 1.29 | 3.60 | 0.46 | 0.93 | 1.65 | 6.99 | 0.56 | 0.95 |
| % Accuracy | −6.39 | 2.24 | 1.23 | 0.32 | −7.67 | 4.48 | 1.72 | −1.67 |
| Recovery (%) | 93.61 | 102.24 | 101.23 | 100.32 | 92.33 | 104.48 | 101.72 | 98.33 |
| Stability Parameter | LQC (3.0) | HQC (2400.0) | LQC (3.0) | HQC (2400.0) | LQC (3.0) | HQC (2400.0) | LQC (3.0) | HQC (2400.0) |
|---|---|---|---|---|---|---|---|---|
| Mean | SD | RSD (%) | Accuracy (%) | |||||
| Freeze–thaw stability (three cycles at −80 °C) | 2.97 | 2457.60 | 0.09 | 80.16 | 2.87 | 3.26 | −0.87 | 2.40 |
| Auto-sampler stability (24 h at 15 °C) | 2.97 | 2405.52 | 0.11 | 45.97 | 3.60 | 1.91 | −1.06 | 0.23 |
| Long-term stability (−80 ˚C for 28 d) | 2.99 | 2484.24 | 0.09 | 69.09 | 3.16 | 2.78 | −0.24 | 3.51 |
| Short-term stability (4 h at room temperature) | 2.96 | 2441.76 | 0.09 | 74.31 | 3.04 | 3.04 | −1.37 | 1.74 |
| Time in Min. | Average a (ng/mL) | X b | ln X | Linearity Parameters |
|---|---|---|---|---|
| 0.0 | 638.34 | 100.00 | 4.61 | Regression equation: y = −0.05287x + 4.649 |
| 2.5 | 587.34 | 92.01 | 4.52 | |
| 5.0 | 521.27 | 81.66 | 4.36 | R2 = 0.9908 |
| 7.5 | 457.24 | 71.63 | 4.14 | |
| 15.0 | 318.15 | 49.84 | 3.97 | Slope: −0.05287 |
| 20.0 | 219.14 | 34.33 | 3.60 | |
| 30.0 | 168.07 | 26.33 | 3.47 | t1/2: 13.11 min |
| 40.0 | 154.10 | 24.14 | 3.34 | Clint: 61.85 mL/min/kg |
| 50.0 | 158.05 | 24.76 | 3.30 | |
| 70.0 | 150.07 | 23.51 | 3.27 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Attwa, M.W.; Abdelhameed, A.S.; Alsibaee, A.M.; Kadi, A.A. A Rapid and Sensitive UPLC-MS/MS Method for Quantifying Capmatinib in Human Liver Microsomes: Evaluation of Metabolic Stability by In Silico and In Vitro Analysis. Separations 2023, 10, 247. https://doi.org/10.3390/separations10040247
Attwa MW, Abdelhameed AS, Alsibaee AM, Kadi AA. A Rapid and Sensitive UPLC-MS/MS Method for Quantifying Capmatinib in Human Liver Microsomes: Evaluation of Metabolic Stability by In Silico and In Vitro Analysis. Separations. 2023; 10(4):247. https://doi.org/10.3390/separations10040247
Chicago/Turabian StyleAttwa, Mohamed W., Ali S. Abdelhameed, Aishah M. Alsibaee, and Adnan A. Kadi. 2023. "A Rapid and Sensitive UPLC-MS/MS Method for Quantifying Capmatinib in Human Liver Microsomes: Evaluation of Metabolic Stability by In Silico and In Vitro Analysis" Separations 10, no. 4: 247. https://doi.org/10.3390/separations10040247
APA StyleAttwa, M. W., Abdelhameed, A. S., Alsibaee, A. M., & Kadi, A. A. (2023). A Rapid and Sensitive UPLC-MS/MS Method for Quantifying Capmatinib in Human Liver Microsomes: Evaluation of Metabolic Stability by In Silico and In Vitro Analysis. Separations, 10(4), 247. https://doi.org/10.3390/separations10040247