Olive Pomace-Derived Biomasses Fractionation through a Two-Step Extraction Based on the Use of Ultrasounds: Chemical Characteristics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents, Standards, and Samples
2.1.1. Reagents and Standards
2.1.2. Samples
2.2. Determination of the Chemical Composition
2.3. Ultrasound-Assisted Extraction
2.4. Total Phenol Content and Antioxidant Capacity Assays
2.5. HPLC-Mass Spectrometry (MS) and Diode Array Analyses
2.6. Alkaline Extraction of the Residual Extraction Fraction and Determination of the Protein Content and Profile
2.7. Sugars Analysis and Lignin Determination in the Alkaline Extracts
2.8. Statistical Analysis
3. Results and Discussion
3.1. Chemical Composition
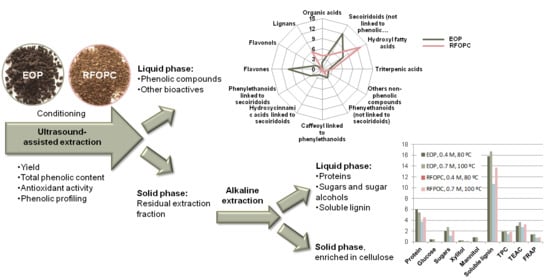
3.2. Extraction of Antioxidants by UAE as a First Valorization Step
3.2.1. Yield, Total Phenolic Content, and Antioxidant Characteristics of the Extracts
3.2.2. Characterization of the Antioxidant Extracts by HPLC-MS Analyses
Phenolic Compounds
Non-Phenolic Compounds
3.3. Second Extraction Step Using Akaline Conditions: Evaluation of the Solubilization of Proteins, Sugars and Lignin
3.3.1. Solubilized Protein
3.3.2. Sugars, Sugar Alcohols, and Lignin
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| EOP | extracted (or exhausted) olive pomace |
| FRAP | ferric ion reducing antioxidant power |
| HPLC | high performance liquid chromatography |
| IT | ion trap |
| MS | mass spectrometry |
| OML | olive mill leaves |
| OL | olive leaves |
| ORAC | oxygen radical absorbance capacity |
| QTOF | quadrupole-time-of-flight |
| RFOPC | residual fraction from olive pit cleaning (or residual pulp) |
| RP | reversed phase |
| TEAC | Trolox equivalent antioxidant capacity |
| TPC | total phenol content |
| UAE | ultrasound-assisted extraction |
References
- Rahmanian, N.; Jafari, S.M.; Wani, T.A. Bioactive profile, dehydration, extraction and application of the bioactive components of olive leaves. Trends Food Sci. Technol. 2015, 42, 150–172. [Google Scholar] [CrossRef]
- Talhaoui, N.; Taamalli, A.; Gómez-Caravaca, A.M.; Fernández-Gutiérrez, A.; Segura-Carretero, A. Phenolic compounds in olive leaves: Analytical determination, biotic and abiotic influence, and health benefits. Food Res. Int. 2015, 77, 92–108. [Google Scholar] [CrossRef]
- EC Comission Implementing Regulation (EU) 2017/2470. Off. J. Eur. Union 2017, 351, 72–201.
- Rodrigues, F.; Pimentel, F.B.; Oliveira, M.B.P.P. Olive by-products: Challenge application in cosmetic industry. Ind. Crops Prod. 2015, 70, 116–124. [Google Scholar] [CrossRef]
- Carnevale, R.; Silvestri, R.; Loffredo, L.; Novo, M.; Cammisotto, V.; Castellani, V.; Bartimoccia, S.; Nocella, C.; Violi, F. Oleuropein, a component of extra virgin olive oil, lowers postprandial glycaemia in healthy subjects. Br. J. Clin. Pharmacol. 2018, 84, 1566–1574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filip, R.; Possemiers, S.; Heyerick, A.; Pinheiro, I.; Raszewski, G.; Davicco, M.J.; Coxam, V. Twelve-month consumption of a polyphenol extract from olive (Olea europaea) in a double blind, randomized trial increases serum total osteocalcin levels and improves serum lipid profiles in postmenopausal women with osteopenia. J. Nutr. Health Aging 2015, 19, 77–86. [Google Scholar] [CrossRef]
- Mosca, A.; Crudele, A.; Smeriglio, A.; Braghini, M.R.; Panera, N.; Comparcola, D.; Alterio, A.; Sartorelli, M.R.; Tozzi, G.; Raponi, M.; et al. Antioxidant activity of Hydroxytyrosol and Vitamin E reduces systemic inflammation in children with paediatric NAFLD. Dig. Liver Dis. 2020, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Huertas, E.; Fonolla, J. Hydroxytyrosol supplementation increases vitamin C levels in vivo. A human volunteer trial. Redox Biol. 2017, 11, 384–389. [Google Scholar] [CrossRef]
- Bañares, C.; Martin, D.; Reglero, G.; Torres, C.F. Protective effect of hydroxytyrosol and rosemary extract in a comparative study of the oxidative stability of Echium oil. Food Chem. 2019, 290, 316–323. [Google Scholar] [CrossRef]
- Hammouda, I.B.; Márquez-Ruiz, G.; Holgado, F.; Sonda, A.; Skalicka-Wozniak, K.; Bouaziz, M. RP-UHPLC-DAD-QTOF-MS as a powerful tool of oleuropein and ligstroside characterization in olive-leaf extract and their contribution to the improved performance of refined olive-pomace oil during heating. J. Agric. Food Chem. 2020, 68, 12039–12047. [Google Scholar] [CrossRef]
- Contreras, M.d.M.; Romero, I.; Moya, M.; Castro, E. Olive-derived biomass as a renewable source of value-added products. Process Biochem. 2020, 97, 43–56. [Google Scholar] [CrossRef]
- Medfai, W.; Contreras, M.d.M.; Lama-Muñoz, A.; Mhamdi, R.; Oueslati, I.; Castro, E. How cultivar and extraction conditions affect antioxidants type and extractability for olive leaves valorization. ACS Sustain. Chem. Eng. 2020, 8, 5107–5118. [Google Scholar] [CrossRef]
- Talhaoui, N.; Gómez-Caravaca, A.M.; León, L.; De La Rosa, R.; Fernández-Gutiérrez, A.; Segura-Carretero, A. from olive fruits to olive oil: Phenolic compound transfer in six different olive cultivars grown under the same agronomical conditions. Int. J. Mol. Sci. 2016, 17, 337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Contreras, M.M.; Lama-Muñoz, A.; Espínola, F.; Moya, M.; Romero, I.; Castro, E. Valorization of olive mill leaves through ultrasound-assisted extraction. Food Chem. 2020, 314, 126218. [Google Scholar] [CrossRef]
- Pavez, I.; Lozano-Sánchez, J.; Borrás-Linares, I.; Nuñez, H.; Robert, P.; Segura-Carretero, A. Obtaining an extract rich in phenolic compounds from olive pomace by pressurized liquid extraction. Molecules 2019, 24, 3108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero, C.; Medina, E.; Mateo, M.A.; Brenes, M. New by-products rich in bioactive substances from the olive oil mill processing. J. Sci. Food Agric. 2018, 98, 225–230. [Google Scholar] [CrossRef] [Green Version]
- Sánchez, J.M.; Pérez Jiménez, J.A.; Díaz Villanueva, M.J.; Serrano, A.; Núñez, N.; Giménez, J.L. New techniques developed to quantify the impurities of olive stone as solid biofuel. Renew. Energy 2015, 78, 566–572. [Google Scholar] [CrossRef]
- Ruiz, E.; Romero-García, J.M.; Romero, I.; Manzanares, P.; Negro, M.J.; Castro, E. Olive-derived biomass as a source of energy and chemicals. Biofuel Bioprod. Biorefin. 2017, 6, 246–256. [Google Scholar] [CrossRef]
- Martínez-Patiño, J.C.; Gómez-Cruz, I.; Romero, I.; Gullón, B.; Ruiz, E.; Brnčićc, M.; Castro, E. Ultrasound-assisted extraction as a first step in a biorefinery strategy for valorisation of extracted olive pomace. Energies 2019, 12, 2679. [Google Scholar] [CrossRef] [Green Version]
- Gómez-Cruz, I.; Cara, C.; Romero, I.; Castro, E.; Gullón, B. Valorisation of exhausted olive pomace by an eco-friendly solvent extraction process of natural antioxidants. Antioxidants 2020, 9, 1010. [Google Scholar] [CrossRef]
- Contreras, M.d.M.; Lama-Muñoz, A.; Gutiérrez-Pérez, J.M.; Espínola, F.; Moya, M.; Romero, I.; Castro, E. Integrated process for sequential extraction of bioactive phenolic compounds and proteins from mill and field olive leaves and effects on the lignocellulosic profile. Foods 2019, 8, 531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lama-Muñoz, A.; Contreras, M.d.M.; Espínola, F.; Moya, M.; Romero, I.; Castro, E. Optimization of oleuropein and luteolin-7-O-glucoside extraction from olive leaves by ultrasound-assisted technology. Energies 2019, 12, 2486. [Google Scholar] [CrossRef] [Green Version]
- Romero-García, J.M.; Lama-Muñoz, A.; Rodríguez-Gutiérrez, G.; Moya, M.; Ruiz, E.; Fernández-Bolaños, J.; Castro, E. Obtaining sugars and natural antioxidants from olive leaves by steam-explosion. Food Chem. 2016, 210, 457–465. [Google Scholar] [CrossRef]
- Sluiter, A.; Hames, B.; Ruiz, R.; Scarlata, C.; Sluiter, J.; Templeton, D.; Nrel, D.C. Determination of Structural Carbohydrates and Lignin in Biomass Determination of Structural Carbohydrates and Lignin in Biomass; NREL/TP-510-42618; National Renewable Energy Laboratory: Golden, CO, USA, 2012. [Google Scholar]
- Ammar, S.; Contreras, M.D.M.; Belguith-Hadrich, O.; Bouaziz, M.; Segura-Carretero, A. New insights into the qualitative phenolic profile of Ficus carica L. fruits and leaves from Tunisia using ultra-high-performance liquid chromatography coupled to quadrupole-time-of-flight mass spectrometry and their antioxidant activity. RSC Adv. 2015, 5, 20035–20050. [Google Scholar] [CrossRef]
- Ammar, S.; Contreras, M.d.M.; Gargouri, B.; Segura-Carretero, A.; Bouaziz, M. RP-HPLC-DAD-ESI-QTOF-MS based metabolic profiling of the potential Olea europaea by-product “wood” and its comparison with leaf counterpart. Phytochem. Anal. 2017, 28, 217–229. [Google Scholar] [CrossRef]
- Martínez-Patiño, J.C.; Ruiz, E.; Romero, I.; Cara, C.; López-Linares, J.C.; Castro, E. Combined acid/alkaline-peroxide pretreatment of olive tree biomass for bioethanol production. Bioresour. Technol. 2017, 239, 326–335. [Google Scholar] [CrossRef]
- Guerra, A.; Ferraz, A.; Cotrim, A.R.; Da Silva, F.T. Polymerization of lignin fragments contained in a model effluent by polyphenoloxidases and horseradish peroxidase/hydrogen peroxide system. Enzyme Microb. Technol. 2000, 26, 315–323. [Google Scholar] [CrossRef]
- Lama-Muñoz, A.; de Contreras, M.M.; Espínola, F.; Moya, M.; Romero, I.; Castro, E. Characterization of the lignocellulosic and sugars composition of different olive leaves cultivars. Food Chem. 2020, 329, 127153. [Google Scholar] [CrossRef]
- Manzanares, P.; Ballesteros, I.; Negro, M.J.; González, A.; Oliva, J.M.; Ballesteros, M. Processing of extracted olive oil pomace residue by hydrothermal or dilute acid pretreatment and enzymatic hydrolysis in a biorefinery context. Renew. Energy 2020, 145, 1235–1245. [Google Scholar] [CrossRef]
- Olmo-García, L.; Polari, J.J.; Li, X.; Bajoub, A.; Fernández-Gutiérrez, A.; Wang, S.C.; Carrasco-Pancorbo, A. Deep insight into the minor fractin of virgin olive oil by using LC-MS and GC-MS multi-class methodologies. Food Chem. 2018, 261, 184–193. [Google Scholar]
- Contreras, M.d.M.; Algieri, F.; Rodriguez-Nogales, A.; Gálvez, J.; Segura-Carretero, A. Phytochemical profiling of anti-inflammatory Lavandula extracts via RP–HPLC–DAD–QTOF–MS and –MS/MS: Assessment of their qualitative and quantitative differences. Electrophoresis 2018, 39, 1284–1293. [Google Scholar] [CrossRef] [PubMed]
- Morreel, K.; Dima, O.; Kim, H.; Lu, F.; Niculaes, C.; Vanholme, R.; Dauwe, R.; Goeminne, G.; Inzé, D.; Messens, E.; et al. Mass spectrometry-based sequencing of lignin oligomers. Plant Physiol. 2010, 153, 1464–1478. [Google Scholar] [CrossRef] [Green Version]
- Olmo-García, L.; Kessler, N.; Neuweger, H.; Wendt, K.; Olmo-Peinado, J.M.; Fernández-Gutiérrez, A.; Baessmann, C.; Carrasco-Pancorbo, A. Unravelling the distribution of secondary metabolites in Olea europaea L.: Exhaustive characterization of eight olive-tree derived matrices by complementary platforms (LC-ESI/APCI-MS and GC-APCI-MS). Molecules 2018, 23, 2419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro, T.B.; Oliveira, A.L.; Costa, C.; Nunes, J.; Vicente, A.A.; Pintado, M. Total and sustainable valorisation of olive pomace using a fractionation approach. Appl. Sci. 2020, 10, 6785. [Google Scholar] [CrossRef]
- Peralbo-Molina, Á.; Priego-Capote, F.; Luque De Castro, M.D. Tentative identification of phenolic compounds in olive pomace extracts using liquid chromatography-tandem mass spectrometry with a quadrupole- quadrupole-time-of-flight mass detector. J. Agric. Food Chem. 2012, 60, 11542–11550. [Google Scholar] [CrossRef] [PubMed]
- Olmo-García, L.; Monasterio, R.P.; Sánchez-Arévalo, C.M.; Fernández-Gutiérrez, A.; Olmo-Peinado, J.M.; Carrasco-Pancorbo, A. Characterization of New Olive Fruit Derived Products Obtained by Means of a Novel Processing Method Involving Stone Removal and Dehydration with Zero Waste Generation. J. Agric. Food Chem. 2019, 67, 9295–9306. [Google Scholar] [CrossRef]
- Strehmel, N.; Böttcher, C.; Schmidt, S.; Scheel, D. Profiling of secondary metabolites in root exudates of Arabidopsis thaliana. Phytochemistry 2014, 108, 35–46. [Google Scholar] [CrossRef]
- Nunes, M.A.; Costa, A.S.G.; Bessada, S.; Santos, J.; Puga, H.; Alves, R.C.; Freitas, V.; Oliveira, M.B.P.P. Olive pomace as a valuable source of bioactive compounds: A study regarding its lipid- and water-soluble components. Sci. Total Environ. 2018, 644, 229–236. [Google Scholar] [CrossRef]
- Lozano-Sánchez, J.; Bendini, A.; Di Lecce, G.; Valli, E.; Gallina Toschi, T.; Segura-Carretero, A. Macro and micro functional components of a spreadable olive by-product (pâté) generated by new concept of two-phase decanter. Eur. J. Lipid Sci. Technol. 2017, 119, 1–9. [Google Scholar] [CrossRef]
- Miersch, O.; Neumerkel, J.; Dippe, M.; Stenzel, I.; Wasternack, C. Hydroxylated jasmonates are commonly occurring metabolites of jasmonic acid and contribute to a partial switch-off in jasmonate signaling. New Phytol. 2008, 177, 114–127. [Google Scholar] [CrossRef]
- Jiménez-Sánchez, C.; Lozano-Sánchez, J.; Rodríguez-Pérez, C.; Segura-Carretero, A.; Fernández-Gutiérrez, A. Comprehensive, untargeted, and qualitative RP-HPLC-ESI-QTOF/MS2 metabolite profiling of green asparagus (Asparagus officinalis). J Food Compos. Anal. 2016, 46, 78–87. [Google Scholar] [CrossRef]
- Peragón, J.; Rufino-Palomares, E.E.; Muñoz-Espada, I.; Reyes-Zurita, F.J.; Lupiáñez, J.A. A new HPLC-MS method for measuring maslinic acid and oleanolic acid in HT29 and HepG2 human cancer cells. Int. J. Mol. Sci. 2015, 16, 21681–21694. [Google Scholar] [CrossRef] [Green Version]
- Kyoung-Rok, K.; Deok-Kun, O. Production of hydroxy fatty acids by microbial fatty acid-hydroxylation enzymes. Biotechnol. Adv. 2013, 31, 1473–1485. [Google Scholar] [CrossRef]
- Srivastava, L.M. Cell wall, cell division and cell growth. In Molecular Cell Biology of the Growth and Differentiation of Plant Cells; Srivastava, L.M., Ed.; Academic Press: San Diego, CA, USA, 2002; pp. 23–74. [Google Scholar]
- Ladaniya, M.S. Fruit Biochemistry. In Citrus Fruit: Biology, Technology and Evaluation; Ladaniya, M., Ed.; Academic Press: San Diego, CA, USA, 2008; pp. 125–190. [Google Scholar]
- Taamalli, A.; Lozano Sánchez, J.; Jebabli, H.; Trabelsi, N.; Abaza, L.; Segura Carretero, A.; Youl Cho, J.; Arráez Román, D. Monitoring the bioactive compounds status in Olea europaea according to collecting period and drying conditions. Energies 2019, 12, 947. [Google Scholar] [CrossRef] [Green Version]
- de la Torre, R.; Carbó, M.; Pujadas, M.; Biel, S.; Mesa, M.D.; Covas, M.I.; Expósito, M.; Espejo, J.A.; Sanchez-Rodriguez, E.; Díaz-Pellicer, P.; et al. Pharmacokinetics of maslinic and oleanolic acids from olive oil–Effects on endothelial function in healthy adults. A randomized, controlled, dose–response study. Food Chem. 2020, 322, 126676. [Google Scholar] [CrossRef] [PubMed]
- Sari, Y.W.; Mulder, W.J.; Sanders, J.P.M.; Bruins, M.E. Towards plant protein refinery: Review on protein extraction using alkali and potential enzymatic assistance. J. Biotechnol. 2015, 10, 1138–1157. [Google Scholar] [CrossRef]
- Contreras, M.d.M.; Lama-Muñoz, A.; Manuel Gutiérrez-Pérez, J.; Espínola, F.; Moya, M.; Castro, E. Protein extraction from agri-food residues for integration in biorefinery: Potential techniques and current status. Bioresour. Technol. 2019, 280, 459–477. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Sanders, J.P.M.; Bruins, M.E. Critical parameters in cost-effective alkaline extraction for high protein yield from leaves. Biomass Bioenergy 2014, 67, 466–472. [Google Scholar] [CrossRef]
- Sari, Y.W.; Syafitri, U.; Sanders, J.P.M.; Bruins, M.E. How biomass composition determines protein extractability. Ind. Crops Prod. 2015, 70, 125–133. [Google Scholar] [CrossRef]
- Montealegre, C.; Esteve, C.; García, M.C.; García-Ruiz, C.; Marina, M.L. Proteins in olive fruit and oil. Crit. Rev. Food Sci. Nutr. 2014, 54, 611–624. [Google Scholar] [CrossRef]
- Lorenzi, V.; Maury, J.; Casanova, J.; Berti, L. Purification, product characterization and kinetic properties of lipoxygenase from olive fruit (Olea europaea L.). Plant Physiol. Biochem. 2006, 44, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Esteve, C.; D’Amato, A.; Marina, M.L.; García, M.C.; Citterio, A.; Righetti, P.G. Identification of olive (Olea europaea) seed and pulp proteins by nLC-MS/MS via combinatorial peptide ligand libraries. J. Proteom. 2012, 75, 2396–2403. [Google Scholar] [CrossRef] [PubMed]
- Vioque, J.; Clemente, A.; Sáchez-Vioque, R.; Pedroche, J.; Millán, F. Effect of alcalase on olive pomace protein extraction. J Am. Oil Chem. Soc. 2000, 77, 181–185. [Google Scholar] [CrossRef]
- McIntosh, S.; Vancov, T. Optimisation of dilute alkaline pretreatment for enzymatic saccharification of wheat straw. Biomass Bioenergy 2011, 35, 3094–3103. [Google Scholar] [CrossRef]
- Rabetafika, H.N.; Bchir, B.; Blecker, C.; Paquot, M.; Wathelet, B. Comparative study of alkaline extraction process of hemicelluloses from pear pomace. Biomass Bioenergy 2014, 61, 254–264. [Google Scholar] [CrossRef]
- Soares, M.L.; Gouveia, E.R. Influence of the alkaline delignification on the simultaneous saccharification and fermentation (SSF) of sugar cane bagasse. Bioresour. Technol. 2013, 147, 645–648. [Google Scholar] [CrossRef]
- Erdocia, X.; Prado, R.; Corcuera, M.Á.; Labidi, J. Effect of different organosolv treatments on the structure and properties of olive tree pruning lignin. J. Ind. Eng. Chem. 2014, 20, 1103–1108. [Google Scholar] [CrossRef]
- Lin, S.Y. Ultraviolet Spectrophotometry. In Methods in Lignin Chemistry; Lin, S.Y., Dence, C.W., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 217–232. [Google Scholar]
- García, A.; Spigno, G.; Labidi, J. Antioxidant and biocide behaviour of lignin fractions from apple tree pruning residues. Ind. Crops Prod. 2017, 104, 242–252. [Google Scholar] [CrossRef]







| Component (%) | OL 4 | OML 4 | EOP | RFOPC |
|---|---|---|---|---|
| Chemical characterization | ||||
| Protein | 9.34 ± 0.35 | 8.10 ± 0.38 | 9.36 ± 0.44 | 4.50 ± 0.33 |
| Glucans 1 | 6.98 ± 0.13 | 9.89 ± 0.57 | 6.96 ± 0.35 | 12.23 ± 0.96 |
| Glucose | 7.68 ± 0.14 | 10.88 ± 0.62 | 7.65 ± 0.38 | 13.45 ± 1.06 |
| Hemicellulose 2 | 5.69 ± 0.11 | 7.90 ± 0.18 | 8.17 ± 0.20 | 14.22 ± 0.99 |
| Galactose | 1.41 ± 0.07 | 1.58 ± 0.07 | 0.38 ± 0.02 | 0.48 ± 0.03 |
| Mannose | 0.60 ± 0.09 | 0.14 ± 0.01 | ND | ND |
| Xylose | 1.30 ± 0.10 | 4.57 ± 0.14 | 8.60 ± 0.23 | 15.20 ± 1.09 |
| Arabinose | 3.06 ± 0.06 | 2.59 ± 0.12 | 0.23 ± 0.02 | 0.37 ± 0.02 |
| Acid soluble lignin | 3.43 ± 0.06 | 2.58 ± 0.026 | 1.65 ± 0.011 | 1.46 ± 0.11 |
| Acid insoluble lignin | 12.66 ± 0.75 | 20.75 ± 0.43 | 19.22 ± 1.63 | 30.79 ± 2.51 |
| Ash | 5.07 ± 0.07 | 10.15 ± 0.10 | 10.06 ± 0.40 | 2.99 ± 0.02 |
| Extractives | 45.07 ± 1.49 | 35.77 ± 1.29 | 52.85 ± 0.72 | 26.29 ± 4.04 |
| Aqueous | 29.46 ± 0.34 | 23.08 ± 1.78 | 48.71 ± 0.54 | 8.39 ± 0.64 |
| Ethanolic | 15.61 ± 1.16 | 12.69 ± 0.51 | 4.14 ± 0.40 | 17.91 ± 3.48 |
| Monomeric sugars 3 | 8.79 | 2.86 ± 0.13 | 5.58 ± 0.04 | 1.54 ± 0.15 |
| Oligomeric sugars 3 | 8.46 ± 0.59 | 5.86 ± 0.68 | 10.27 ± 1.09 | |
| Glucose 3 | 7.33 ± 0.17 | 7.61 ± 0.60 | 7.90 ± 0.36 | 1.31 ± 0.43 |
| Galactose 3 | 0.94 ± 0.03 | 1.36 ± 0.09 | 1.19 ± 0.07 | 1.93 ± 0.19 |
| Mannose 3 | ND | 0.03 ± 0.02 | 0.42 ± 0.03 | 0.46 ± 0.07 |
| Xylose 3 | 0.11 ± 0.01 | 0.31 ± 0.13 | 0.29 ± 0.26 | 4.97 ± 0.35 |
| Arabinose 3 | 0.41 ± 0.04 | 2.00 ± 0.44 | 1.64 ± 0.08 | 3.13 ± 0.34 |
| Mannitol 3 | 4.64 ± 0.23 | 2.63 ± 0.13 | 5.36 ± 0.07 | 0.64 ± 0.19 |
| Fat | ND | 9.85 ± 0.10 | 2.47 ± 0.34 | 8.72 ± 0.96 |
| Acetyl groups | 0.66 ± 0.01 | 0.77 ± 0.02 | 0.79 ± 0.59 | 1.97 ± 0.06 |
| Elemental analysis | ||||
| N | 1.49 ± 0.06 | 1.30 ± 0.06 | 1.50 ± 0.07 | 0.72 ± 0.05 |
| C | 48.38 ± 0.48 | 48.08 ± 0.66 | 49.65 ± 0.85 | 56.86 ± 1.24 |
| H | 6.48 ± 0.15 | 6.49 ± 0.08 | 6.23 ± 0.33 | 7.26 ± 0.40 |
| Biomass | Extracted Volume/Total Volume | Total Extraction Yield | Solubilization 1 | Extract Richness 2 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TPC 3 | TEAC 3 | FRAP 3 | ORAC 3 | TPC 3 | TEAC 3 | FRAP 3 | ORAC 3 | |||
| OL 4 | 83.33 ± 1.44 c | 24.00 ± 1.99 b | 3.33 ± 0.42 b | 11.03 ± 0.20 b | 14.24 ± 0.78 b | 72.69 ± 3.65 b | 14.03 ± 2.99 a | 46.18 ± 4.44 b | 59.53 ± 4.48 a | 272.77 ± 33.35 a |
| OML 4 | 83.33 ± 0.00 c | 15.77 ± 0.88 c | 2.06 ± 0.06 c | 11.94 ± 2.96 b | ND | ND | 13.11 ± 0.68 a | 74.72 ± 19.33 a | ND | ND |
| EOP | 92.17 ± 0.00 a | 37.44 ± 0.38 a | 4.45 ± 0.09 a | 25.25 ± 0.20 a | 20.30 ± 1.28 a | 95.30 ± 5.93 a | 11.90 ± 0.30 a | 67.43 ± 1.08 a | 54.23 ± 3.62 a | 254.44 ± 13.78 a |
| RFOPC | 85.80 ± 0.25 b | 6.70 ± 0.33 d | 0.46 ± 0.02 d | 2.61 ± 0.31 c | 1.35 ± 0.16 c | 11.50 ± 0.87 c | 6.92 ± 0.33 b | 39.14 ± 6.09 b | 20.10 ± 1.83 b | 171.78 ± 12.98 b |
| N° | RT 1 (Min) | Molecular Formula 2 | m/z2,3 | Error (Ppm) 3 | Score 2 | MS/MS 1 | Compound | Class | OL | OML | EOP | RFOPC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1.2 | C14H20O8 | 315.11 | −3.4 | 94 | 153, 135, 123 | Hydroxytyrosol glucoside | Phenylethanoids (not linked to secoiridoids) | + | − | + | + |
| 2 | 1.1 | C8H10O3 | 153.06 | 1.1 | 99 | 123 | Hydroxytyrosol | Phenylethanoids (not linked to secoiridoids) | + | + | + | + |
| 3 | 1.9 | C14H20O7 | 299.11 | 0.5 | 95 | 179, 161, 119, 101 | Tyrosol glucoside | Phenylethanoids (not linked to secoiridoids) | − | − | + | − |
| 4 | 7.3 | C27H30O15 | 593.15 | −2.8 | 94 | 503, 473, 383, 353 | Apigenin 6,8-di-C-glucoside | Flavones | + | + | − | − |
| 5 | 8.1 | C27H30O16 | 609.15 | −3.2 | 90 | 447, 285 | Luteolin di-hexoxide 1 | Flavone | + | + | − | − |
| 6 | 8.4 | C25H32O14 | 555.17 | −3.5 | 91 | 537, 403, 323, 223 | Hydroxyoleuropein 1 | Phenylethanoids (linked to secoiridoids) | + | + | − | − |
| 7 | 8.4 | C27H30O16 | 609.15 | −2.7 | 94 | 447, 285 | Luteolin di-hexoside 2 | Flavone | + | + | − | − |
| 8 | 8.5 | C24H30O13 | 525.16 | −3.4 | 91 | 481, 389, 319, 195, 165 | Demethyloleuropein | Phenylethanoids (linked to secoiridoids) | + | + | − | − |
| 9 | 8.9 | C25H32O15 | 571.17 | −3.5 | 92 | 523, 403, 359, 223, 179 | Dihydroxyoleuropein | Phenylethanoids (linked to secoiridoids) | + | − | − | − |
| 10 | 9.2 | C27H30O16 | 609.15 | −3.5 | 91 | 447, 285 | Luteolin di-hexoxide 3 | Flavones | + | + | − | − |
| 11 | 9.3 | C25H32O14 | 555.17 | −3.1 | 92 | 537, 403, 371, 323, 223 | Hydroxyoleuropein 2 | Phenylethanoids (linked to secoiridoids) | + | + | − | − |
| 12 | 9.3 | C23H32O11 | 483.19 | −3.5 | 93 | 347, 123 | 3,4-DHPEA-EDA derivative (+ hexose + H2) | Phenylethanoids (linked to secoiridoids) | − | − | + | − |
| 13 | 9.7 | C20H32O12 | 463.18 | −3.0 | 95 | 347, 301 | Quercetin glucoside | Flavonols | + | − | + | − |
| 14 | 9.9 | C27H30O16 | 609.15 | −3.2 | 92 | 447, 301, 179 | Quercetin 3-O-rutinoside | Flavonols | + | + | + | − |
| 15 | 9.9 | C29H36O15 | 623.20 | −3.3 | 91 | 461, 315 | Verbascoside | Caffeoyl phenylethanoid derivatives | + | + | + | − |
| 16 | 10.0 | C21H20O11 | 447.10 | −3.7 | 91 | 285 | Luteolin 7-O-glucoside | Flavones | + | + | + | − |
| 17 | 10.2 | C27H30O15 | 593.15 | −3.6 | 90 | 285 | Luteolin hexoside deoxyhexoside 1 | Flavones | + | + | + | − |
| 18 | 10.5 | C27H30O15 | 593.15 | −3.6 | 92 | 447, 285 | Luteolin hexoside deoxyhexoside 2 | Flavones | + | + | + | − |
| 19 | 10.6 | C31H42O18 | 701.23 | −1.9 | 96 | 539, 377, 307, 275 | Oleuropein hexoside 1 | Phenylethanoids (linked to secoiridoids) | + | + | + | − |
| 20 | 10.7 | C29H36O15 | 623.20 | −3.7 | 90 | 461 | Isoverbascoside | Caffeoyl phenylethanoid derivatives | + | + | + | − |
| 21 | 10.8 | C17H20O7 | 335.11 | −4.0 | 93 | 317, 199, 153, 111 | Hydroxyde (carboxymethyl) oleuropein aglycone | Phenylethanoids (linked to secoiridoids) | − | − | + | − |
| 22 | 11.1 | C31H42O18 | 701.23 | −2.4 | 93 | 539, 437, 377, 307, 275 | Oleuropein hexoside 2 | Phenylethanoids (linked to secoiridoids) | + | + | − | − |
| 23 | 11.2 | C30H34O11 | 569.20 | −2.6 | 95 | 551, 539, 393, 373, 177, 162 | G(8-O-4)S(8-5)G 5 (−C) | Lignan derivatives | − | − | − | + |
| 24 | 11.3 | C27H30O14 | 557.16 | −3.7 | 90 | 269 | Apigenin 7-O-rutinoside | Flavones | + | + | − | − |
| 25 | 11.4 | C25H32O13 | 539.18 | −4.6 | 85 | 403, 223 | Oleouropein 1 | Phenylethanoids (linked to secoiridoids) | − | − | + | − |
| 26 | 11.5 | C21H20O11 | 447.09 | −3.5 | 91 | 285 | Luteolin 7-O-hexoside 1 | Flavones | + | + | − | − |
| 27 | 11.6 | C25H32O13 | 539.18 | −3.1 | 92 | 403, 377, 307, 275, 223 | Oleuropein | Phenylethanoids (linked to secoiridoids) | + | + | + | + |
| 28 | 11.7 | C25H28O14 | 551.14 | −3.6 | 92 | 507, 389, 341, 281, 251, 179, 161 | Caffeoyl-6′-secologanoside | Hydroxycinnamics (linked to secoiridoids) | − | − | + | − |
| 29 | 12.0 | C22H22O11 | 461.11 | −4.2 | 90 | 446, 299, 284 | Diosmetin 7-O-glucoside | Flavones | + | + | − | − |
| 30 | 12.3 | C21H20O11 | 447.09 | −3.5 | 92 | 285 | Luteolin 7-O-hexoside isomer 2 | Flavones | + | + | − | − |
| 31 | 12.4 | C25H32O13 | 539.18 | −2.9 | 92 | 377, 307, 275, 223 | Oleouropein 2 | Phenylethanoids (linked to secoiridoids) | + | + | + | − |
| 32 | 12.7 | C25H32O13 | 539.18 | −3.5 | 92 | 403, 377, 307, 275, 223 | Oleouropein 3 | Phenylethanoids (linked to secoiridoids) | + | + | + | − |
| 33 | 13.0 | C27H36O14 | 583.20 | −2.1 | 95 | 537, 403, 223, 179 | Lucidumoside C | Phenylethanoids (linked to secoiridoids) | + | + | − | − |
| 34 | 13.1 | C17H20O6 | 319.12 | −3.6 | 94 | 183, 181, 153, 111 | 3,4-DHPEA-EDA | Phenylethanoids (linked to secoiridoids) | − | − | + | − |
| 35 | 13.2 | C19H22O8 | 377.13 | −4.3 | 91 | 307, 275 | Oleuropein aglycone 1 | Phenylethanoids (linked to secoiridoids) | + | − | − | − |
| 36 | 13.2 | C25H28O13 | 535.15 | −4.0 | 90 | 491, 389, 345, 265, 163 | p-Coumaroyl-6′-secologanoside | Hydroxycinnamics (linked to secoiridoids) | − | − | + | + |
| 37 | 13.5 | C25H32O12 | 523.18 | −2.9 | 94 | 361, 291, 259, 223 | Ligustroside | Phenylethanoids (linked to secoiridoids) | + | + | + | − |
| 38 | 14.1 | C19H22O8 | 377.13 | −4.0 | 91 | 307, 275 | Oleuropein aglycone 2 | Phenylethanoids (linked to secoiridoids) | + | − | − | − |
| 39 | 14.4 | C21H20O6 | 367.12 | −3.7 | 95 | 352, 337, 336, 322, 307, 177, 162 | S(8-5)G 4 derivative 1 (−H2O) | Lignan derivatives | − | − | − | + |
| 40 | 14.6 | C20H18O5 | 337.11 | −3.9 | 93 | 322, 307, 291, 177, 162 | S(8-5)G 4 derivative 2 (−H2O, −CH2O) | Lignan derivatives | − | − | − | + |
| 41 | 15.0 | C28H34O13 | 577.19 | 4.4 | 92 | 531, 415, 398, 285, 273, 239 | (+)-1-Acetoxypinoresinol 4′-b-O-glucoside | Lignan derivatives | + | − | − | − |
| 42 | 15.4 | C31H28O14 | 623.14 | −2.2 | 94 | 323, 299, 285 | Diosmetin di-hexoside | Flavones | + | + | − | − |
| 43 | 15.4 | C42H54O23 | 925.30 | 0.9 | 97 | 539, 377, 307, 275 | Jaspolyoside isomer 1 | Phenylethanoids (linked to secoiridoids) | + | − | − | − |
| 44 | 15.8 | C31H36O11 | 583.22 | −3.2 | 93 | 565, 535, 413, 387, 373, 357, 343, 195, 165 | G(8-O-4)S(8-8)G 5 | Lignan derivatives | − | − | − | + |
| 45 | 16.2 | C31H34O11 | 581.20 | −2.9 | 93 | 563, 551, 533, 503, 385, 367, 355, 337, 336, 218, 195, 177, 165 | G(8-O-4)S(8-5)G 4 1 | Lignan derivatives | − | − | − | + |
| 46 | 16.4 | C41H50O21 | 877.28 | 0.2 | 99 | 715, 701, 539, 377, 307, 275, 149 | Oleuropein derivative 1 (oleuropein hexoside + C10H8O3) | Phenylethanoids (linked to secoiridoids) | + | + | − | − |
| 47 | 16.9 | C42H54O23 | 925.30 | 0.6 | 96 | 539, 377, 307, 275 | Jaspolyoside isomer 2 | Phenylethanoids (linked to secoiridoids) | + | − | − | − |
| 48 | 17.0 | C31H34O11 | 581.20 | −2.6 | 94 | 551, 367, 355, 337, 218, 195, 165 | G(8-O-4)S(8-5)G 4 2 | Lignan derivatives | − | − | − | + |
| 49 | 17.0 | C34H38O15 | 685.21 | −1.7 | 92 | 539, 377, 307, 275 | Oleuropein derivative 2 (oleuropein + C9H6O2) | Phenylethanoids (linked to secoiridoids) | + | − | − | − |
| 50 | 18.3 | C41H58O20 | 869.34 | 0.6 | 95 | 829, 707, 539, 377, 325, 307, 275, 145 | Oleuropein derivative 3 (oleuropein hexoside + C10H16O2) | Phenylethanoids (linked to secoiridoids) | + | − | − | − |
| 51 | 18.7 | C19H22O7 | 361.13 | −1.6 | 98 | 329, 291, 225, 193, 181 | Hydroxytyrosol linked to desoxy elenolic acid | Phenylethanoids (linked to secoiridoids) | − | − | + | − |
| 52 | 19.3 | C35H48O15 | 707.29 | −0.5 | 97 | 539, 377, 307, 275 | Oleuropein derivative 4 (oleuropein + C10H16O2) | Phenylethanoids (linked to secoiridoids) | + | − | − | − |
| 53 | 26.2 | C43H60O14 | 799.39 | 0.04 | 99 | 539, 377, 307, 277 | Oleuropein derivative 6 (oleuropein + C18H28O) | Phenylethanoids (linked to secoiridoids) | + | − | − | − |
| 54 | 26.8 | C41H62O14 | 777.41 | 0.23 | 98 | 539, 377, 307, 275 | Oleuropein derivative 7 (oleuropein + C16H30O) | Phenylethanoids (linked to secoiridoids) | + | − | − | − |
| 55 | 14.2 | C15H10O6 | 285.04 | −2.12 | 98 | 175, 151 | Luteolin | Flavones | − | + | + | − |
| No | RT 1 (Min) | Molecular Formula 2 | m/z2,3 | Error 3 (Ppm) | Score 3 | MS/MS Fragments 1 | Compound | OL | OML | EOP | RFOPC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1′ | 0.4 | C6H14O6 | 181.072 | -0.2 | 99 | 163, 119 | Mannitol | + | + | + | + |
| 2′ | 0.5 | C6H8O7 | 191.020 | −0.7 | 99 | 173, 111 | Citric acid | + | + | + | + |
| 3′ | 0.5 | C7H12O6 | 191.057 | −2.1 | 99 | Quinic acid | + | + | + | + | |
| 4′ | 0.5 | C6H12O7 | 195.052 | −2.9 | 98 | 177, 129 | Gluconic acid | + | + | − | + |
| 5′ | 1.6 | C17H28O11 | 407.158 | −4.5 | 90 | 389, 375, 357, 313, 151 | Loganin derivative (+H2; +O) | + | − | + | − |
| 6′ | 3.7 | C12H18O7S | 305.071 | −3.9 | 94 | 225, 97 | 12−Hydroxyjasmonic acid sulfate | + | + | − | − |
| 7′ | 2.5 | C16H22O11 | 389.110 | −2.8 | 93 | 345, 302, 209, 165, 139, 121 | Oleoside/Secologanoside | + | + | + | + |
| 8′ | 3.3 | C9H12O4 | 183.066 | 0.8 | 99 | 139 | Oleoside/secologanoside derivative (-glucosyl; −CO2) or decarboxymethylelenolic acid | − | + | + | + |
| 9′ | 3.9 | C16H22O11 | 389.111 | −4 | 91 | 345, 302, 209, 187, 165, 139, 121 | Oleoside/Secologanoside | − | − | + | − |
| 10′ | 4.4 | C18H28O12 | 435.153 | −4.1 | 90 | 389, 357, 313, 151 | Loganin derivative (+H2; +O; +CO) | − | − | + | − |
| 11′ | 4.7 | C17H24O11 | 403.13 | −4.3 | 90 | 371, 223, 179 | Elenolic acid hexoside 1 | + | − | − | − |
| 12′ | 5.3 | C12H20O7S | 307.09 | −4.4 | 92 | 227, 165, 97 | Dihydrohydroxyjasmonic acid sulfate derivative | − | − | + | − |
| 13′ | 5.3 | C10H14O4 | 197.08 | 0.7 | 98 | 153 | Elenolic acid hexoside derivative (+H2; -CO; -Hexose) | − | − | − | + |
| 14′ | 5.5 | C16H26O10 | 377.15 | −4.4 | 90 | 197, 153 | Elenolic acid hexoside derivative (+H2; -CO) | + | + | + | + |
| 15′ | 6.2 | C17H26O11 | 405.14 | −4.7 | 90 | 373, 181 | Elenolic acid hexoside derivative (+H2) | − | − | + | − |
| 16′ | 6.5 | C17H24O11 | 403.13 | −4 | 92 | 371, 223, 179 | Elenolic acid hexoside 2 | + | + | + | − |
| 17′ | 6.9 | C20H34O13 | 481.19 | −3.9 | 90 | 371, 151 | Unknown (elenolic acid hexoside derivative) | − | − | + | − |
| 18′ | 7.5 | C11H14O7 | 257.07 | −1 | 99 | 239, 225, 195, 137 | Hydroxyelenolic acid | − | − | + | + |
| 19′ | 7.8 | C17H24O11 | 403.13 | −4.2 | 92 | 241, 223 | Elenolic acid hexoside 3 | + | + | + | − |
| 20′ | 9.3 | C9H16O4 | 187.1 | −1 | 99 | 125 | Azelaic acid | − | + | − | + |
| 21′ | 11.6 | C18H34O6 | 345.23 | −2.5 | 96 | 201, 171 | Tetrahydroxyoctadecenoic acid isomer 1 | − | − | − | + |
| 22′ | 12.1 | C18H34O6 | 345.23 | −3.4 | 95 | 327, 309, 201, 171 | Tetrahydroxyoctadecenoic acid isomer 2 | − | − | − | + |
| 23′ | 12.4 | C18H34O6 | 345.23 | −4.2 | 91 | 327, 309, 201 | Tetrahydroxyoctadecenoic acid isomer 3 | − | − | − | + |
| 24′ | 15.5 | C26H38O13 | 557.23 | 3.6 | 91 | 513, 345, 227, 185 | 6′-O-[(2E)-2,6-Dimethyl-8-hydroxy-2-octenoyloxy]-secologanoside | + | + | + | − |
| 25′ | 15.8 | C18H32O5 | 327.22 | −3.8 | 93 | 201, 171 | Trihydroxyoctadecadienoic acid | − | − | − | + |
| 26′ | 16.4 | C18H32O6 | 343.21 | −3.8 | 93 | 325, 307, 245, 201 | Dihydroxyoctadecenedioic acid | − | − | − | + |
| 27′ | 16.7 | C18H34O5 | 329.23 | −2.4 | 95 | 201 | Trihydroxyoctadecenoic acid | − | − | + | + |
| 28′ | 17.7 | C18H34O6 | 345.23 | −4.2 | 91 | 327, 309, 265, 247 | Dihydroxyoctadecanedioic acid | − | − | − | + |
| 29′ | 18.1 | C18H36O5 | 331.25 | −4 | 92 | 312.7 | Trihydroxyoctadecanoic acid | − | + | + | + |
| 30′ | 18.9 | C16H32O4 | 287.22 | −3.3 | 95 | 269, 210 | Dihydroxyhexadecanoic acid | + | + | + | + |
| 31′ | 21.7 | C30H48O5 | 487.34 | −3.8 | 90 | 467 | Hydroxylated derivative of maslinic acid | − | − | − | + |
| 32′ | 21.2 | C20H36O6 | 371.25 | −3.4 | 93 | 329, 311, 201 | Trihydroxyoctadecenoic acid + C2H2O (acetyl) | − | − | + | + |
| 33′ | 24.7 | C36H64O9 | 639.45 | −2.9 | 93 | 329, 327, 201 | Dimer formed by trihydroxyoctadecadienoic acid and trihydroxyoctadecenoic acid | − | − | + | + |
| 34′ | 24.9 | C36H66O9 | 641.47 | −2.3 | 93 | 329, 201 | Di-trihydroxyoctadecenoic acid | − | − | + | + |
| 35′ | 25.1 | C30H48O4 | 471.35 | −3.4 | 94 | 453, 407 | Pomolic acid | + | + | − | − |
| 36′ | 25.3 | C34H64O8 | 599.45 | −2.7 | 94 | 329, 287, 201 | Dimer formed by trihydroxyoctadecenoic acid and dihydroxyhexadecanoic acid | − | − | + | + |
| 37′ | 25.7 | C30H48O4 | 471.35 | −3.3 | 93 | 423, 405 | Maslinic acid | + | + | + | + |
| 38′ | 26 | C18H36O3 | 299.26 | −2.9 | 95 | 281, 253 | Hydroxyoctadecanoic acid | − | − | − | + |
| 39′ | 26.9 | C30H48O3 | 455.35 | −3.4 | 93 | 407, 395 | Oleanolic acid | + | + | + | + |
| Byproduct 1 | Mild | Strong | Mild | Strong |
|---|---|---|---|---|
| Solubilized Protein (g/L) | Solubilized Protein (g/L) | Recovery (%) | Recovery (%) | |
| OML 2 | 3.9 ± 0.1 b | 5.1 ± 0.5 a | 48.7 ± 1.5 c | 63.1 ± 5.7 b |
| OL 2 | ND | 5.2 ± 0.4 | ND | 55.5 ± 4.3 b |
| EOP | 6.0 ± 0.3 a | 5.4 ± 0.4 a | 63.9 ± 2.3 b | 58.2 ± 3.3 b |
| RFOPC | 3.7 ± 0.3 b | 4.5 ± 0.2 a | 81.3 ± 5.8 a | 100.1 ± 3.0 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Contreras, M.d.M.; Gómez-Cruz, I.; Romero, I.; Castro, E. Olive Pomace-Derived Biomasses Fractionation through a Two-Step Extraction Based on the Use of Ultrasounds: Chemical Characteristics. Foods 2021, 10, 111. https://doi.org/10.3390/foods10010111
Contreras MdM, Gómez-Cruz I, Romero I, Castro E. Olive Pomace-Derived Biomasses Fractionation through a Two-Step Extraction Based on the Use of Ultrasounds: Chemical Characteristics. Foods. 2021; 10(1):111. https://doi.org/10.3390/foods10010111
Chicago/Turabian StyleContreras, María del Mar, Irene Gómez-Cruz, Inmaculada Romero, and Eulogio Castro. 2021. "Olive Pomace-Derived Biomasses Fractionation through a Two-Step Extraction Based on the Use of Ultrasounds: Chemical Characteristics" Foods 10, no. 1: 111. https://doi.org/10.3390/foods10010111
APA StyleContreras, M. d. M., Gómez-Cruz, I., Romero, I., & Castro, E. (2021). Olive Pomace-Derived Biomasses Fractionation through a Two-Step Extraction Based on the Use of Ultrasounds: Chemical Characteristics. Foods, 10(1), 111. https://doi.org/10.3390/foods10010111