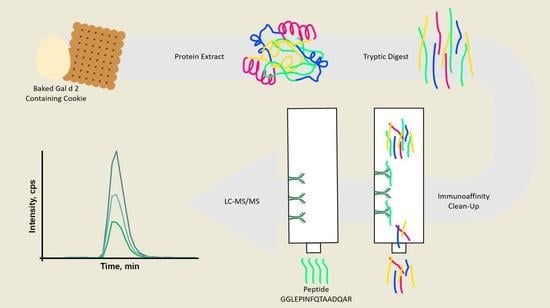
Improved Sensitivity of Allergen Detection by Immunoaffinity LC-MS/MS Using Ovalbumin as a Case Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Selection of Suitable Tryptic Peptides for Gal d 2
2.2. Generation of Monoclonal Antibodies
2.3. Screening Hybridoma Supernatants by Indirect Gal d 2 ELISA
2.4. Screening Hybridoma Supernatants by Indirect Peptide ELISA
2.5. Preparation and Characterization of Processed Material Containing Ovalbumin
2.6. Western Blot
2.7. Surface Plasmon Resonance (SPR) Spectroscopy
2.8. Coupling of Monoclonal Antibodies to the Affinity Matrix
2.9. Protein Extraction for Antibody Affinity Matrix Clean-Up
2.10. Tryptic Digestion of Proteins
2.11. Immunoaffinity Clean-Up of Gal d 2 or Tryptic Gal d 2 Peptides
2.12. Solid-Phase Extraction (SPE)
2.13. LC-MS/MS
3. Results
3.1. Selection of Peptides and Corresponding Peptide Specific Monoclonal Antibodies
3.2. Western Blot
3.3. Characterization of Monoclonal Antibodies by SPR Spectroscopy
3.4. Commercial Sandwich ELISA (Morinaga)
3.5. Immuno-Affinity LC-MS/MS Analysis (Clean-Up after Tryptic Digestion)
3.6. Immuno-Affinity LC-MS/MS Analysis (Clean-Up after Extraction)
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- van Hengel, A.J. Food allergen detection methods and the challenge to protect food-allergic consumers. Anal. Bioanal. Chem. 2007, 389, 111–118. [Google Scholar] [CrossRef]
- López-Pedrouso, M.; Lorenzo, J.M.; Gagaoua, M.; Franco, D. Current trends in proteomic advances for food allergen analysis. Biology 2020, 9, 247. [Google Scholar] [CrossRef]
- Cucu, T.; Jacxsens, L.; de Meulenaer, B. Analysis to support allergen risk management: Which way to go? J. Agric. Food Chem. 2013, 61, 5624–5633. [Google Scholar] [CrossRef]
- Köppel, R.; Dvorak, V.; Zimmerli, F.; Breitenmoser, A.; Eugster, A.; Waiblinger, H.-U. Two tetraplex real-time PCR for the detection and quantification of DNA from eight allergens in food. Eur. Food Res. Technol. 2010, 230, 367–374. [Google Scholar] [CrossRef]
- Walker, M.J.; Burns, D.T.; Elliott, C.T.; Gowland, M.H.; Mills, E.N.C. Is food allergen analysis flawed? Health and supply chain risks and a proposed framework to address urgent analytical needs. Analyst 2016, 141, 24–35. [Google Scholar] [CrossRef] [Green Version]
- Holzhauser, T.; Johnson, P.; Hindley, J.P.; O’Connor, G.; Chan, C.-H.; Costa, J.; Fæste, C.K.; Hirst, B.J.; Lambertini, F.; Miani, M.; et al. Are current analytical methods suitable to verify VITAL® 2.0/3.0 allergen reference doses for EU allergens in foods? Food Chem. Toxicol. 2020, 145, 111709. [Google Scholar] [CrossRef] [PubMed]
- Croote, D.; Quake, S.R. Food allergen detection by mass spectrometry: The role of systems biology. NPJ Syst. Biol. Appl. 2016, 2, 16022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allergen Nomenclature: WHO/IUIS Allergen Nomenclature Sub-Committee. Available online: http://www.allergen.org/viewallergen.php?aid=336 (accessed on 26 November 2021).
- Becker, J.O.; Hoofnagle, A.N. Replacing immunoassays with tryptic digestion-peptide immunoaffinity enrichment and LC-MS/MS. Bioanalysis 2012, 4, 281–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khuda, S.; Slate, A.; Pereira, M.; Al-Taher, F.; Jackson, L.; Diaz-Amigo, C.; Bigley, E.C.; Whitaker, T.; Williams, K.M. Effect of processing on recovery and variability associated with immunochemical analytical methods for multiple allergens in a single matrix: Sugar cookies. J. Agric. Food Chem. 2012, 60, 4195–4203. [Google Scholar] [CrossRef]
- Knudsen, J.; Otte, J.; Olsen, K.; Skibsted, L. Effect of high hydrostatic pressure on the conformation of β-lactoglobulin A as assessed by proteolytic peptide profiling. Int. Dairy J. 2002, 12, 791–803. [Google Scholar] [CrossRef]
- Naderi, N.; House, J.D.; Pouliot, Y.; Doyen, A. Effects of high hydrostatic pressure processing on hen egg compounds and egg products. Compr. Rev. Food Sci. Food Saf. 2017, 16, 707–720. [Google Scholar] [CrossRef] [Green Version]
- Parker, C.H.; Khuda, S.E.; Pereira, M.; Ross, M.M.; Fu, T.-J.; Fan, X.; Wu, Y.; Williams, K.M.; DeVries, J.; Pulvermacher, B.; et al. Multi-Allergen quantitation and the impact of thermal treatment in industry-processed baked goods by ELISA and liquid chromatography-tandem mass spectrometry. J. Agric. Food Chem. 2015, 63, 10669–10680. [Google Scholar] [CrossRef] [PubMed]
- Azarnia, S.; Boye, J.I.; Mongeon, V.; Sabik, H. Detection of ovalbumin in egg white, whole egg and incurred pasta using LC–ESI-MS/MS and ELISA. Food Res. Int. 2013, 52, 526–534. [Google Scholar] [CrossRef]
- Gavage, M.; van Vlierberghe, K.; van Poucke, C.; de Loose, M.; Gevaert, K.; Dieu, M.; Renard, P.; Arnould, T.; Gillard, N. Selection of egg peptide biomarkers in processed food products by high resolution mass spectrometry. J. Chromatogr. A 2019, 1584, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Korte, R.; Oberleitner, D.; Brockmeyer, J. Determination of food allergens by LC-MS: Impacts of sample preparation, food matrix, and thermal processing on peptide detectability and quantification. J. Proteom. 2019, 196, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Monaci, L.; Losito, I.; de Angelis, E.; Pilolli, R.; Visconti, A. Multi-allergen quantification of fining-related egg and milk proteins in white wines by high-resolution mass spectrometry. Rapid Commun. Mass Spectrom. 2013, 27, 2009–2018. [Google Scholar] [CrossRef]
- Yamasaki, M.; Takahashi, N.; Hirose, M. Crystal structure of S-ovalbumin as a non-loop-inserted thermostabilized serpin form. J. Biol. Chem. 2003, 278, 35524–35530. [Google Scholar] [CrossRef] [Green Version]
- Ueberham, E.; Spiegel, H.; Havenith, H.; Rautenberger, P.; Lidzba, N.; Schillberg, S.; Lehmann, J. Simplified tracking of a soy allergen in processed food using a monoclonal antibody-based sandwich ELISA targeting the soybean 2S albumin Gly m 8. J. Agric. Food Chem. 2019, 67, 8660–8667. [Google Scholar] [CrossRef] [Green Version]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- O’Farrell, P.H. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 1975, 250, 4007–4021. [Google Scholar] [CrossRef]
- de Jong, W.W.; Zweers, A.; Cohen, L.H. Influence of single amino acid substitutions on electrophoretic mobility of sodium dodecyl sulfate-protein complexes. Biochem. Biophys. Res. Commun. 1978, 82, 532–539. [Google Scholar] [CrossRef]
- Rath, A.; Glibowicka, M.; Nadeau, V.G.; Chen, G.; Deber, C.M. Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc. Natl. Acad. Sci. USA 2009, 106, 1760–1765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aquino, A.; Conte-Junior, C.A. A systematic review of food allergy: Nanobiosensor and food allergen detection. Biosensors 2020, 10, 194. [Google Scholar] [CrossRef]
- Nimata, M.; Okada, H.; Kurihara, K.; Sugimoto, T.; Honjoh, T.; Kuroda, K.; Yano, T.; Tachibana, H.; Shoji, M. A harmonized immunoassay with liquid chromatography-mass spectrometry analysis in egg allergen determination. Anal. Bioanal. Chem. 2018, 410, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Mattarozzi, M.; Careri, M. The role of incurred materials in method development and validation to account for food processing effects in food allergen analysis. Anal. Bioanal. Chem. 2019, 411, 4465–4480. [Google Scholar] [CrossRef]
- Török, K.; Hajas, L.; Horváth, V.; Schall, E.; Bugyi, Z.; Kemény, S.; Tömösközi, S. Identification of the factors affecting the analytical results of food allergen ELISA methods. Eur. Food Res. Technol. 2015, 241, 127–136. [Google Scholar] [CrossRef]
- Holm, B.E.; Bergmann, A.C.; Hansen, P.R.; Koch, C.; Houen, G.; Trier, N.H. Antibodies with specificity for native and denatured forms of ovalbumin differ in reactivity between enzyme-linked immunosorbent assays. APMIS Acta Pathol. Microbiol. Immunol. Scand. 2015, 123, 136–145. [Google Scholar] [CrossRef]
- Walczyk, N.E.; Smith, P.M.C.; Tovey, E.R.; Roberts, T.H. Peanut protein extraction conditions strongly influence yield of allergens Ara h 1 and 2 and sensitivity of immunoassays. Food Chem. 2017, 221, 335–344. [Google Scholar] [CrossRef]
- Ito, K.; Yamamoto, T.; Oyama, Y.; Tsuruma, R.; Saito, E.; Saito, Y.; Ozu, T.; Honjoh, T.; Adachi, R.; Sakai, S.; et al. Food allergen analysis for processed food using a novel extraction method to eliminate harmful reagents for both ELISA and lateral-flow tests. Anal. Bioanal. Chem. 2016, 408, 5973–5984. [Google Scholar] [CrossRef]
- Lidzba, N.; García Arteaga, V.; Schiermeyer, A.; Havenith, H.; Muranyi, I.; Schillberg, S.; Lehmann, J.; Ueberham, E. Development of monoclonal antibodies against pea globulins for multiplex assays targeting legume proteins. J. Agric. Food Chem. 2021, 69, 2864–2874. [Google Scholar] [CrossRef]
- Schubert-Ullrich, P.; Rudolf, J.; Ansari, P.; Galler, B.; Führer, M.; Molinelli, A.; Baumgartner, S. Commercialized rapid immunoanalytical tests for determination of allergenic food proteins: An overview. Anal. Bioanal. Chem. 2009, 395, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Fu, T.-J.; Maks, N.; Banaszewski, K. Effect of heat treatment on the quantitative detection of egg protein residues by commercial enzyme-linked immunosorbent assay test kits. J. Agric. Food Chem. 2010, 58, 4831–4838. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, S.; Xu, C.; Caperna, T.J.; Garrett, W.M. Comparison of protein solubilization methods suitable for proteomic analysis of soybean seed proteins. Anal. Biochem. 2005, 342, 214–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monaci, L.; de Angelis, E.; Guagnano, R.; Ganci, A.P.; Garaguso, I.; Fiocchi, A.; Pilolli, R. Validation of a ms based proteomics method for milk and egg quantification in cookies at the lowest VITAL levels: An alternative to the use of precautionary labeling. Foods 2020, 9, 1489. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Parker, C.H.; Boo, C.C.; Fiedler, K.L. Comparison of allergen quantification strategies for egg, milk, and peanut in food using targeted LC-MS/MS. Anal. Bioanal. Chem. 2021, 413, 5755–5766. [Google Scholar] [CrossRef]








| Position of Cleavage Site | Peptide Sequence | Peptide Length [aa] | Peptide Mass [kDa] |
|---|---|---|---|
| 17 | MGSIGAASMEFCFDVFK | 17 | 1840.157 |
| 20 | ELK | 3 | 388.464 |
| 47 | VHHANENIFYCPIAIMSALAMVYLGAK | 27 | 2977.55 |
| 51 | DSTR | 4 | 477.475 |
| 56 | TQINK | 5 | 602.688 |
| 59 | VVR | 3 | 372.468 |
| 62 | FDK | 3 | 408.455 |
| 85 | LPGFGDSIEAQCGTSVNVHSSLR | 23 | 2374.61 |
| 105 | DILNQITKPNDVYSFSLASR | 20 | 2281.55 |
| 111 | LYAEER | 6 | 779.848 |
| 123 | YPILPEYLQCVK | 12 | 1465.771 |
| 127 | ELYR | 4 | 579.654 |
| 143 | GGLEPINFQTAADQAR | 16 | 1687.829 |
| 159 | ELINSWVESQTNGIIR | 16 | 1859.069 |
| 182 | NVLQPSSVDSQTAMVLVNAIVFK | 23 | 2460.871 |
| 187 | GLWEK | 5 | 631.729 |
| 190 | AFK | 3 | 364.445 |
| 200 | DEDTQAMPFR | 10 | 1209.296 |
| 219 | VTEQESKPVQMMYQIGLFR | 19 | 2284.674 |
| 227 | VASMASEK | 8 | 821.944 |
| 229 | MK | 2 | 277.382 |
| 264 | ILELPFASGTMSMLVLLPDEVSGLEQLESIINFEK | 35 | 3864.521 |
| 277 | LTEWTSSNVMEER | 13 | 1581.717 |
| 278 | K | 1 | 146.189 |
| 280 | IK | 2 | 259.349 |
| 285 | VYLPR | 5 | 646.787 |
| 287 | MK | 2 | 277.382 |
| 291 | MEEK | 4 | 535.613 |
| 323 | YNLTSVLMAMGITDVFSSSANLSGISSAESLK | 32 | 3294.736 |
| 340 | ISQAVHAAHAEINEAGR | 17 | 1773.926 |
| 360 | EVVGSAEAGVDAASVSEEFR | 20 | 2009.114 |
| 370 | ADHPFLFCIK | 10 | 1190.425 |
| 382 | HIATNAVLFFGR | 12 | 1345.567 |
| 386 | CVSP | 4 | 404.482 |
| Q1 (m/z) | (Q3 m/z) | RT (min) | Marker Peptide | DP (V) | CE (V) | CXP (V) |
|---|---|---|---|---|---|---|
| 761.1 | 930.5 | 6.85 | Gal d 2 DILNQITKPNDVYSFSLASR. + 3y8 | 141 | 45 | 26 |
| 761.1 | 767.4 | 6.85 | Gal d 2 DILNQITKPNDVYSFSLASR. + 3y7 | 141 | 31 | 22 |
| 930.0 | 1116.6 | 6.93 | Gal d 2 ELINSWVESQTNGIIR. + 2y10 | 161 | 45 | 32 |
| 620.3 | 673.4 | 6.93 | Gal d 2 ELINSWVESQTNGIIR. + 3y6 | 141 | 25 | 20 |
| 620.3 | 572.4 | 6.93 | Gal d 2 ELINSWVESQTNGIIR. + 3y5 | 171 | 25 | 42 |
| 844.4 | 1121.5 | 6.12 | Gal d 2 GGLEPINFQTAADQAR. + 2y10 | 156 | 43 | 34 |
| 844.4 | 666.3 | 6.12 | Gal d 2 GGLEPINFQTAADQAR. + 2y12 + 2 | 151 | 35 | 20 |
| 563.3 | 560.3 | 6.12 | Gal d 2 GGLEPINFQTAADQAR. + 3y5 | 76 | 17 | 40 |
| 761.9 | 1036.5 | 6.74 | Gal d 2 YPILPEYLQCVK. + 2y8 | 151 | 35 | 30 |
| 761.9 | 518.8 | 6.74 | Gal d 2 YPILPEYLQCVK. + 2y8 + 2 | 151 | 29 | 30 |
| 673.4 | 1024.6 | 6.24 | Gal d2 HIATNAVLFFGR.2y9 | 146 | 35 | 28 |
| 673.4 | 923.5 | 6.24 | Gal d2 HIATNAVLFFGR. + 2y8 | 146 | 35 | 26 |
| 673.4 | 809.5 | 6.20 | Gal d2 HIATNAVLFFGR. + 2y7 | 16 | 37 | 26 |
| 791.4 | 1052.5 | 5.75 | Gal d2 LTEWTSSNVMEER 2y9 | 146 | 37 | 32 |
| 791.4 | 951.4 | 5.75 | Gal d2 LTEWTSSNVMEER 2y8 | 141 | 37 | 28 |
| 887.5 | 1138.6 | 4.42 | Gal d2 ISQAVHAAHAEINEAGR. + 2y11 | 151 | 59 | 38 |
| 887.5 | 1067.5 | 4.42 | Gal d2 ISQAVHAAHAEINEAGR. + 2y10 | 151 | 53 | 36 |
| 887.5 | 996.5 | 4.42 | Gal d2 ISQAVHAAHAEINEAGR. + 2y9 | 161 | 53 | 32 |
| 624.3 | 924.5 | 6.10 | Gal d2 ADHPFLFCIK. + 2y7 | 61 | 39 | 30 |
| 624.3 | 827.4 | 6.10 | Gal d2 ADHPFLFCIK. + 2y6 | 71 | 35 | 28 |
| 624.3 | 324.1 | 6.10 | Gal d2 ADHPFLFCIK. + 2b3 | 11 | 29 | 10 |
| 624.3 | 681.3 | 6.10 | Gal d2 ADHPFLFCIK. + 2b6 | 66 | 35 | 24 |
| Premix Added to Dough * [g/kg] | Calculated ppm Ovalbumin in Dough ** | Quantified ppm Ovalbumin by ELISA in Cookies |
|---|---|---|
| 0 | - | 0 |
| 1.7 | 3.1 | 1.1 ± 0.01 |
| 3.3 | 6.2 | 2.6 ± 0.12 |
| 10.0 | 18.6 | 11.7 ± 0.32 |
| 31.0 | 57.7 | 49.6 ± 2.19 |
| 61.8 | 113.3 | 89.6 ± 2.75 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Röder, M.; Wiacek, C.; Lankamp, F.; Kreyer, J.; Weber, W.; Ueberham, E. Improved Sensitivity of Allergen Detection by Immunoaffinity LC-MS/MS Using Ovalbumin as a Case Study. Foods 2021, 10, 2932. https://doi.org/10.3390/foods10122932
Röder M, Wiacek C, Lankamp F, Kreyer J, Weber W, Ueberham E. Improved Sensitivity of Allergen Detection by Immunoaffinity LC-MS/MS Using Ovalbumin as a Case Study. Foods. 2021; 10(12):2932. https://doi.org/10.3390/foods10122932
Chicago/Turabian StyleRöder, Martin, Claudia Wiacek, Frauke Lankamp, Jonathan Kreyer, Wolfgang Weber, and Elke Ueberham. 2021. "Improved Sensitivity of Allergen Detection by Immunoaffinity LC-MS/MS Using Ovalbumin as a Case Study" Foods 10, no. 12: 2932. https://doi.org/10.3390/foods10122932
APA StyleRöder, M., Wiacek, C., Lankamp, F., Kreyer, J., Weber, W., & Ueberham, E. (2021). Improved Sensitivity of Allergen Detection by Immunoaffinity LC-MS/MS Using Ovalbumin as a Case Study. Foods, 10(12), 2932. https://doi.org/10.3390/foods10122932