Modernization of Control of Pathogenic Micro-Organisms in the Food-Chain Requires a Durable Role for Immunoaffinity-Based Detection Methodology—A Review
Abstract
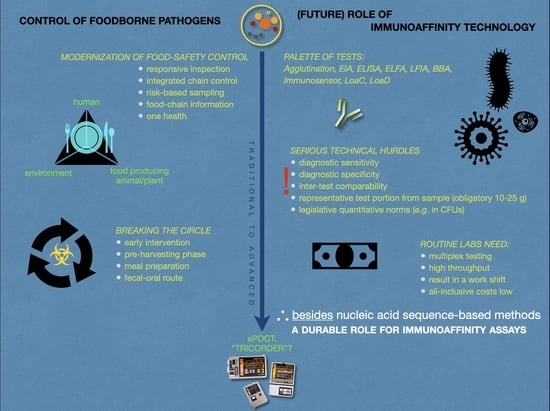
:1. Introduction
1.1. Food and Micro-Organisms
1.2. Need to Contain Foodborne Pathogenic Micro-Organisms
1.2.1. Bacteria
1.2.2. Parasites
1.2.3. Viruses
1.2.4. Other Types of Pathogens
1.3. Failing Containment of Pathogenic Micro-Organisms
2. Analytical Microbiology
2.1. Why Need to Measure?
2.2. Historical Overview of the Discovery of Micro-Organisms
2.3. Immunoaffinity Principle
2.3.1. Microbial Handles for Analysis
2.3.2. Antigens and Antibodies as Potential Analytical Tools
2.3.2.1. Measuring Principle of Direct Antigen Tests
2.3.2.2. Measuring Principle of Indirect Antigen Tests
- (i)
- Low immunogenic response of the individual animal, and
- (ii)
- The so-called seroconversion window.
2.3.3. Sample Type and Preparation
2.3.3.1. Sample Preparation for Direct Antigen Tests
2.3.3.2. Sample Preparation for Indirect Antigen Tests
2.4. Where and How to Measure?
- (i)
- The investigator is adequately educated and trained to perform the analysis.
- (ii)
- The place where the test is performed is appropriate and hygienic measures are adequate and not a source for false positive (or false-negative) results.
- (iii)
- The test is fit for its intended purpose.
- (1)
- Asymptomatic carriers of a pathogen that remain unnoticed and are not excluded from the food-chain or not further investigated following a visual sanitary inspection.
- (2)
- The sometimes extremely low microbial dose causing disease in humans which therefore needs very sensitive analytical methods, and
- (3)
- the overwhelming presence of many other, non-harmful, entities obscuring the detection of a disease-causative agent.
2.5. Test What, When, for Which Purpose and at What Costs?
- (1)
- Which microbes need to be analyzed and intervened?
- (2)
- Where in the food-chain can these MOs best analyzed and with how many of which (type of) samples? (see also above)
- (3)
- What are the test quality requirements?
- (4)
- What is the most effective test methodology?
2.6. Immunoaffinity Assays
2.6.1. Traditional Methods
2.6.1.1. Direct Antigen Approaches
2.6.1.2. Indirect Antigen Approaches
2.6.2. Advanced Methods
2.6.2.1. Bead-Based Arrays
2.6.2.2. Immunosensors
2.6.2.3. Microfluidic Devices
2.6.3. Concluding Remarks on IA Assays
2.7. Available Test Principles Other Than Immunoaffinity
2.7.1. Bacteriophages
2.7.2. Nucleic Acid
2.7.3. Physicochemical Approaches
3. Reliability of Results
3.1. Method Validation
- Sample: Host-MO interactions affecting the composition of the sample and its analyte concentration. Surprisingly, this can thus be different from one geographical region to another.
- Assay system: Physical, chemical, biological, managerial, acclimation, housing, and technician-related factors affecting the capacity of the assay to detect a specific analyte. Here, sources of errors are not necessarily random and independent. They are for example laboratory effects, method bias, matrix variation effects, random and systemic errors of measurement, run effects, or bias.
- Other sources of errors that affect the capacity of a test result to predict accurately the contamination status of animals, food products, plants, or populations relative to the analyte in question.
3.2. Predictive Values
4. Example: Salmonella Detection in The Pork-Production Chain
5. General Discussion and Conclusions
5.1. Responsive and Smart Monitoring and Control
5.2. Prediction, Indicators, and Prevention of Sherlock’s Holmes Statistics
5.3. The Weak Link
5.4. Fool’s Gold?
5.5. Validity and Comparability of Results
5.6. Weighing the Investment in New Methodology
5.7. Bioprepology
5.8. Conclusions and Messages
- -
- A great part of the ante and postmortem monitoring comprises indirect antigen assays gauging specific antibodies in serum, meat juice, and oral fluids.
- -
- When (intracellularly) hidden or low body burden, IA assays outperform other analytical techniques, including NASB methods.
- -
- IA assays offer a chief advantage over NASB assays: they can detect acellular biomolecules, including toxins, uncovering a (past) infection.
- -
- The largest part of analyses worldwide involves the ante- and post-mortem monitoring of MOs in the (pre-)harvesting phase of the food-chain. Almost 50% of all tests involve measuring Salmonella. IMS plays an important role.
- -
- Whatever analytical sensitivity, analytical specificity, and other test characteristics, the applied assay should fit the purpose while it is clear when and where it is used in the food-chain.
- -
- Novel methods should be presented with data from field samples, not from spiked or polished reference samples exclusively.
- -
- Integrated chain control and One Health principles in combination with risk-based sampling are imperative to combat effectively current and (re-)emerging pathogens while increasing the safety level of food.
- -
- Successful intervention on the guidance of (environmental) monitoring will also protect (families of) farmers and food-workers as a good health and safety practice.
- -
- The need for more speed and sensitivity is modest and not prominent in field laboratories, albeit results within a working shift are highly desired.
- -
- Mutual comparison of results produced by a gamut of alternative analysis systems and comparison with reference methods is an unsolved challenge.
- -
- In the case of group assessment, routine laboratories prefer high diagnostic specificity, multiplexing, and high throughput, but convincing low all-inclusive costs even more.
- -
- All steps between the decision to sample and conversion of a sample into a test portion need continuous and careful attention or the analysis result becomes less reliable or even worthless. Sampling, sample treatment, and sample processing are of cardinal importance.
- -
- In spite of the numerous innovative techniques that evolved over the last decades, only a few have been authorized for screening, monitoring, and control programs.
- -
- The food analysis field is conservative for several understandable reasons, not only because of financial risks. Routine laboratories are bound to accreditation and providing results as if generated by reference methods.
6. Future Perspectives
- (1)
- Test performances that are compliant with local, national, and international (ISO) quality standards.
- (2)
- Enabling the use of (relatively) easily available, preferably non-invasive, samples, such as egg, feces, hair/feathers, saliva/mucus, urine.
- (3)
- Able to analyze simultaneously different types of analytes, such as cells, oligo-/polynucleotides, oligo-/polypeptides, organic metabolites, toxins, in a single run.
- (4)
- Easy-to-use or automated platform demanding minimal user involvement.
- (5)
- Giving accurate results instantaneously (i.e., within 10 min in a PoC situation or otherwise in a work shift).
- (6)
- At low costs, while,
- (7)
- Using a robust, reliable portable multiplex point-of-care testing device (xPOCT) and reagents that have a long shelf-life at ambient temperatures, and which are easily disposed of after use.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AB | antibody |
| AC | (relative) accuracy |
| AFNOR | Association Française de Normalisation |
| AG | antigen |
| AgBP | antigen-binding protein |
| AOAC-RI | Association of Official Agricultural Chemists Research Institute |
| ATP | adenosine triphosphate |
| BBA | bead-based assay |
| BSE | bovine spongiform encephalopathy |
| CE | Conformité Européenne (standard mark) |
| CFU | colony-forming units |
| CPA | cross-priming amplification |
| CPS | capsular polysaccharides |
| CSF | classic swine fever |
| CT | cycle threshold (as in qPCR) |
| DALY | disability-adjusted life year |
| DES | diethylstilbestrol |
| DIVA | differentiating vaccinated from infected animals |
| EF | extracellular factor (of Streptococcus suis) |
| EFSA | European Food Safety Authority |
| EIA | enzyme immunoassay |
| ELFA | enzyme-linked immunofluorescent assay |
| ELISA | enzyme-linked immunosorbent assay |
| ESI | electrospray ionization |
| FCI | food-chain information |
| FSO | food-safety objective |
| GC | gas-chromatography |
| GAP | good agricultural practice |
| GHP | good hygiene practice |
| GMP | good manufacturing practice |
| GVP | good veterinary practice |
| HACCP | hazard analysis critical control points |
| HAV | hepatitis A virus |
| HEV | hepatitis E virus |
| HRP | horseradish peroxidase |
| IA | immunoaffinity |
| ICT | immunochromatographic test |
| IG | immunoglobulin |
| IgA | immunoglobulin class A |
| IgG | immunoglobulin class G |
| IgM | immunoglobulin class M |
| IMS | immunomagnetic separation |
| iPCR | immuno-PCR |
| iqPCR | real-time immunoquantative PCR |
| IR | infra-red |
| ISO | International Organization for Standardization |
| LAMP | loop-mediated isothermal amplification |
| LFIA | lateral flow immunoassay |
| LIMS | laboratory information management system |
| LFD | lateral flow device |
| LoaC | lab-on-a-chip |
| LoaD | lab-on-a-disc |
| LOD | limit-of-detection |
| LPS | lipopolysaccharides |
| MAB | monoclonal antibody |
| MALDI-TOF | matrix-assisted laser-desorption ionization time-of-flight |
| MIP | molecular imprinted polymer |
| MO | micro-organism |
| MS | mass spectrometry |
| NASB | nucleic acid sequence-based |
| NASBA | nucleic acid sequence-based amplification |
| NGO | non-governmental organization |
| NMR | nuclear magnetic resonance |
| NPV | negative predictive value |
| OD | optical density |
| OH | one health |
| OIE | Office International des Epizooties (World Organization for Animal Health) |
| PAB | polyclonal antibody |
| PFGE | pulsed-field gel electrophoresis |
| PoC | point-of-care |
| ppb | parts per billion |
| PRRS | porcine reproduction and respiratory syndrome |
| PRV | pseudorabies virus |
| qPCR | quantitative (real-time) polymerase chain reaction |
| RCA | rolling circle amplification |
| RPA | recombinase polymerase amplification |
| RT | reverse transcription |
| RT-PCR | reverse transcription polymerase chain reaction |
| SDA | strand displacement amplification |
| SE | (relative) sensitivity |
| SERS | surface-enhanced Raman-spectroscopy |
| SP | (relative) specificity |
| SPR | surface plasmon resonance |
| STEC | Shiga toxin-producing Escherichia coli |
| TTR | time-to-result |
| VBNC | viable but non-culturable |
| UKAS | United Kingdom Accreditation Service |
| VOC | volatile organic compound |
| WHO | World Health Organization |
| xPOCT | multiplexed point-of-care testing |
References
- Martyushev, L.M.; Seleznev, V.D. Maximum entropy production principle in physics, chemistry and biology. Phys. Rep. 2006, 426, 1–45. [Google Scholar] [CrossRef]
- Silva, C.; Annamalai, K. Entropy Generation and Human Aging: Lifespan Entropy and Effect of Physical Activity Level. Entropy 2008, 10, 100–123. [Google Scholar] [CrossRef] [Green Version]
- Commission of The European Communities. Commission Regulation (EC) No 2073/2005 of 15 November 2005 on microbiological criteria for foodstuffs (consolidated version including Commission Regulation amendments and corrections). Off. J. Eur. Union 2005, L338, 1–26. [Google Scholar]
- Janssen, R.; Krogfelt, K.A.; Cawthraw, S.A.; Van Pelt, W.; Wagenaar, J.A.; Owen, R.J. Host-pathogen interactions in Campylobacter infections: The host perspective. Clin. Microbiol. Rev. 2008, 21, 505–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishu Allos, B.; Iovine, N.M.; Blaser, M.J. Campylobacter jejuni and Related Species. In Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases, 8th ed.; Bennett, J.E., Dolin, R., Blaser, M.J., Eds.; W.B. Saunders: Philadelphia, PA, USA, 2015; Chapter 2018; Volume 2, pp. 2485–2493.e4. [Google Scholar] [CrossRef]
- Mangen, M.J.J.; Havelaar, A.H.; de Wit, G.A. Campylobacteriosis and Sequelae in the Netherlands—Estimating the Disease Burden and the Cost-of-Illness, National Institute for Public Health and the Environment, Centre for Prevention and Health Services Research (RIVM), RIVM Report 250911004/2004. Available online: https://www.rivm.nl/bibliotheek/rapporten/250911004.pdf (accessed on 16 December 2020).
- Overgaauw, P.A.M. Parasite risks from raw meat-based diets for companion animals. Companion Anim. 2020, 25, 261–267. [Google Scholar] [CrossRef]
- Carter, M.E.; Quinn, P.J. Salmonella infections in dogs and cats. In Salmonella in Domestic Animals; Wray, C., Wray, A., Eds.; CABI Publishing: Wallingford, UK, 2000; pp. 231–244. [Google Scholar]
- Nemser, S.M.; Doran, T.; Grabenstein, M.; McConnell, T.; McGrath, T.; Pamboukian, R.; Smith, A.C.; Achen, M.; Danzeisen, G.; Kim, S.; et al. Investigation of Listeria, Salmonella, and toxigenic Escherichia coli in various pet foods. Foodborne Pathog. Dis. 2014, 11, 706–709. [Google Scholar] [CrossRef] [Green Version]
- Peeples, E.H., Jr. Meanwhile, Humans Eat Pet Food. The New York Times. 16 December 1975, p. 39. Available online: https://www.nytimes.com/1975/12/16/archives/meanwhile-humans-eat-pet-food.html (accessed on 17 December 2020).
- Greig, J.D.; Todd, E.C.D.; Bartleson, C.A.; Michaels, B.S. Outbreaks where food workers have been implicated in the spread of foodborne disease. Part 1. Description of the problem, methods, and agents involved. J. Food Prot. 2007, 70, 1752–1761. [Google Scholar] [CrossRef] [Green Version]
- Battelli, G. Zoonoses as occupational diseases. Vet. Ital. 2008, 44, 601–609. [Google Scholar] [PubMed]
- Rabozzi, G.; Bonizzi, L.; Crespi, E.; Somaruga, C.; Sokooti, M.; Tabibi, R.; Vellere, F.; Brambilla, G.; Colosio, C. Emerging zoonoses: The “One Health approach”. Saf. Health Work 2012, 3, 77–83. [Google Scholar] [CrossRef] [Green Version]
- Food Safety. Available online: https://www.who.int/news-room/fact-sheets/detail/food-safety (accessed on 2 December 2020).
- Lanata, C.F.; Fischer-Walker, C.L.; Olascoaga, A.C.; Torres, C.X.; Aryee, M.J.; Black, R.E. Global causes of diarrheal disease mortality in children <5 years of age: A systematic review. PLoS ONE 2013, 8, e72788. [Google Scholar] [CrossRef] [Green Version]
- European Food Safety Authority and European Centre for Disease Prevention and Control. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 2018, 16, 1–262. [Google Scholar] [CrossRef]
- Kirk, M.D.; Pires, S.M.; Black, R.E.; Caipo, M.; Crump, J.A.; Devleesschauwer, B.; Döpfer, D.; Fazil, A.; Fischer-Walker, C.L.; Hald, T.; et al. World Health Organization Estimates of the Global and Regional Disease Burden of 22 Foodborne Bacterial, Protozoal, and Viral Diseases, 2010: A Data Synthesis. PLoS Med. 2015, 12, e1001921. [Google Scholar] [CrossRef] [Green Version]
- European Food Safety Authority and European Centre for Disease Prevention and Control. The European Union One Health 2018 Zoonoses Report. EFSA J. 2019, 17, 1–276. [Google Scholar] [CrossRef] [Green Version]
- European Food Safety Authority and European Centre for Disease Prevention and Control. The European Union One Health 2019 Zoonoses Report. EFSA J. 2021, 19, 6406. [Google Scholar] [CrossRef]
- Foley, S.L.; Lynne, A.M.; Nayak, R. Salmonella challenges: Prevalence in swine and poultry and potential pathogenicity of such isolates. J. Anim. Sci. 2008, 86, E149–E162. [Google Scholar] [CrossRef]
- Healy, B.; Cooney, S.; O’Brien, S.; Iversen, C.; Whyte, P.; Nally, J.; Callanan, J.J.; Fanning, S. Cronobacter (Enterobacter sakazakii): An opportunistic foodborne pathogen. Foodborne Pathog. Dis. 2010, 7, 339–350. [Google Scholar] [CrossRef]
- Fusco, V.; Abriouel, H.; Benomar, N.; Kabisch, J.; Chieffi, D.; Cho, G.-S.; Franz, C.M.A.P. Opportunistic Food-Borne Pathogens. In Food Safety and Preservation; Grumezescu, A.M., Holban, A.M., Eds.; Academic Press: London, UK, 2018; pp. 269–306. [Google Scholar] [CrossRef]
- Lipman, L.J.A.; Van Knapen, F. Voedselinfecties en intoxicaties. In Inleiding tot de Levensmiddelenhygiëne—Achtergrond en Feiten, 2nd ed.; Lipman, L.J.A., Ruiter, A., Eds.; Reed Business: Amsterdam, The Netherlands, 2011; pp. 89–114. (In Dutch) [Google Scholar]
- Roberts, T.; Murrell, K.D.; Marks, S. Economic losses caused by foodborne parasitic diseases. J. Parasitol. 1994, 10, 419–423. [Google Scholar] [CrossRef] [Green Version]
- Chengat Prakashbabu, B.; Marshall, L.R.; Crotta, M.; Gilbert, W.; Johnson, J.C.; Alban, L.; Guitian, J. Risk-based inspection as a cost-effective strategy to reduce human exposure to cysticerci of Taenia saginata in low-prevalence settings. Parasites Vectors 2018, 11, 257. [Google Scholar] [CrossRef] [Green Version]
- Laranjo-González, M.; Devleesschauwer, B.; Gabriël, S.; Dorny, P.; Allepuz, A. Epidemiology, impact and control of bovine cysticercosis in Europe: A systematic review. Parasites Vectors 2016, 9, 81. [Google Scholar] [CrossRef] [Green Version]
- Jansen, F.; Dorny, P.; Berkvens, D.; Van Hul, A.; Van den Broeck, N.; Makay, C.; Praet, N.; Gabriël, S. Assessment of the repeatability and border-plate effects of the B158/B60 enzyme-linked-immunosorbent assay for the detection of circulating antigens (Ag-ELISA) of Taenia saginata. Vet. Parasitol. 2016, 227, 69–72. [Google Scholar] [CrossRef]
- Regional Office for Europe of the World Health Organization. The Burden of Foodborne Diseases in the WHO European Region; Kruse, H., Ed.; Copenhagen, Denmark, 2017. Available online: https://www.euro.who.int/__data/assets/pdf_file/0005/402989/50607-WHO-Food-Safety-publicationV4_Web.pdf (accessed on 22 January 2021).
- Teunis, P.F.M.; Le Guyader, F.S.; Liu, P.; Ollivier, J.; Moe, C.L. Noroviruses are highly infectious but there is strong variation in host susceptibility and virus pathogenicity. Epidemics 2020, 32, 100401. [Google Scholar] [CrossRef] [PubMed]
- Potasman, I.; Paz, A.; Odeh, M. Infectious outbreaks associated with bivalve shellfish consumption: A worldwide perspective. Clin. Infect. Dis. 2002, 35, 921–928. [Google Scholar] [CrossRef] [Green Version]
- European Food Safety Authority Panel on Biological Hazards; Ricci, A.; Allende, A.; Bolton, D.; Chemaly, M.; Davies, R.; Fernandez Escamez, P.S.; Herman, L.; Koutsoumanis, K.; Lindqvist, R.; et al. Scientific Opinion on the public health risks associated with hepatitis E virus (HEV) as a food-borne pathogen. EFSA J. 2017, 15, 4886. [Google Scholar] [CrossRef]
- Gofflot, S.; El Moualij, B.; Zorzi, D.; Melen, L.; Roels, S.; Quatpers, D.; Grassi, J.; Vanopdenbosch, E.; Heinen, E.; Zorzi, W. Immuno-Quantitative Polymerase Chain Reaction for Detection and Quantitation of Prion Protein. J. Immunoassay Immunochem. 2004, 25, 241–258. [Google Scholar] [CrossRef] [PubMed]
- Eskola, M.; Kos, G.; Elliott, C.T.; Hajšlová, J.; Mayar, S.; Krska, R. Worldwide contamination of food-crops with mycotoxins: Validity of the widely cited ‘FAO estimate’ of 25%, Critical Reviews in Food Science and Nutrition. Crit. Rev. Food Sci. Nutr. 2020, 60, 2773–2789. [Google Scholar] [CrossRef] [PubMed]
- Noor, R. Insight to Foodborne Diseases: Proposed Models for Infections and Intoxications. Biomed. Biotechnol. Res. J. 2019, 3, 135–139. [Google Scholar] [CrossRef]
- International Society for Infectious Diseases, ProMed. Available online: https://promedmail.org (accessed on 28 January 2021).
- Ferreira, V.; Wiedmann, M.; Teixeira, P.; Stasiewicz, M.J. Listeria monocytogenes Persistence in Food-Associated Environments: Epidemiology, Strain Characteristics, and Implications for Public Health. J. Food Prot. 2014, 77, 150–170. [Google Scholar] [CrossRef] [PubMed]
- Azizoglu, R.O.; Osborne, J.; Wilson, S.; Kathariou, S. Role of Growth Temperature in Freeze-Thaw Tolerance of Listeria spp. Appl. Environ. Microbiol. 2009, 75, 5315–5320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thévenot, D.; Dernburg, A.; Vernozy-Rozand, C. An updated review of Listeria monocytogenes in the pork meat industry and its products. J. Appl. Microbiol. 2006, 101, 7–17. [Google Scholar] [CrossRef]
- Osimani, A.; Aquilanti, L.; Clementi, F. Salmonellosis associated with mass catering: A survey of European Union cases over a 15-year period. Epidemiol. Infect. 2016, 144, 3000–3012. [Google Scholar] [CrossRef]
- Hugas, M.; Beloeil, P.A. Controlling Salmonella along the in the European Union—Progress over the last ten years. Euro Surveill. 2014, 19, 20804. [Google Scholar] [CrossRef] [Green Version]
- Beumer, R.R.; Kusumaningrum, H. Kitchen hygiene in daily life. Int. Biodeterior. Biodegrad. 2003, 51, 299–302. [Google Scholar] [CrossRef]
- Anderson, J.B.; Shuster, T.A.; Hansen, K.E.; Levy, A.S.; Volk, A. A camera’s view of consumer food-handling behaviors. J. Am. Diet. Assoc. 2004, 104, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Burke, T.; Young, I.; Papadopoulos, A. Assessing food safety knowledge and preferred information sources among 19–29 year olds. Food Control 2016, 69, 83–89. [Google Scholar] [CrossRef]
- Koch, S.; Lohmann, M.; Geppert, J.; Stamminger, R.; Epp, A.; Böl, G.-F. Kitchen Hygiene in the Spotlight: How Cooking Shows Influence Viewers’ Hygiene Practices. Risk Anal. 2020, 41, 131–140. [Google Scholar] [CrossRef]
- Satin, M. Death in the Pot: The Impact of Food Poisoning on History, 2nd ed.; Prometheus Books: New York, NY, USA, 2009; p. 258. [Google Scholar]
- Dixon, B. Power Unseen: How Microbes Rule the World; W.H. Freeman & Company Limited: Oxford, UK, 1994; p. 237. [Google Scholar]
- Satin, M. History Foodborne Disease—Part I—Ancient History. In Encyclopedia of Food Safety, 1st ed.; Motarjemi, Y., Moy, G., Todd, E., Eds.; Volume 1 History, Science and Methods; Academic Press: London, UK, 2014; p. 2304. [Google Scholar]
- Foster, E.M. Historical overview of key issues in food safety. Emerg. Infect. Dis. 1997, 3, 481–482. [Google Scholar] [CrossRef]
- Stephany, R.W. Hormonal growth promoting agents in food producing animals. In Handbook of Experimental Pharmacology; Thieme, D., Hemmersbach, P., Eds.; Doping in Sports; Springer: Berlin/Heidelberg, Germany, 2010; Volume 195, pp. 355–367. [Google Scholar] [CrossRef]
- Gribble, G.W. Food chemistry and chemophobia. Food Sec. 2013, 5, 177–187. [Google Scholar] [CrossRef]
- Hameed, S.; Xie, L.; Ying, Y. Conventional and emerging detection techniques for pathogenic bacteria in food science: A review. Trends Food Sci. Technol. 2018, 81, 61–73. [Google Scholar] [CrossRef]
- Saravanan, A.; Senthil Kumar, P.; Hemavathy, R.V.; Jeevanantham, S.; Kamalesh, R.; Sneha, S.; Yaashikaa, P.R. Methods of detection of food-borne pathogens: A review. Environ. Chem. Lett. 2021, 19, 189–207. [Google Scholar] [CrossRef]
- Visansirikul, S.; Kolodziej, S.A.; Demchenko, A.V. Staphylococcus aureus capsular polysaccharides: A structural and synthetic perspective. Org. Biomol. Chem. 2020, 18, 783. [Google Scholar] [CrossRef]
- Vinogradov, E.; Conlan, J.W.; Perry, M.B. Serological cross-reaction between the lipopolysaccharide O-polysaccharide antigens of Escherichia coli O157:H7 and strains of Citrobacter freundii and Citrobacter sedlakii. FEMS Microbiol. Lett. 1998, 190, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Gal-Mor, O.; Boyle, E.C.; Grassl, G.A. Same species, different diseases: How and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Front. Microbiol. 2014, 5, 391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paoli, G.C.; Kleina, L.G.; Brewster, J.D. Development of Listeria monocytogenes–specific immunomagnetic beads using a single-chain antibody fragment. Foodborne Pathog. Dis. 2007, 4, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Gaastra, W. Micro-organismen. In Inleiding tot de Levensmiddelenhygiëne—Achtergrond en Feiten, 2nd ed.; Lipman, L.J.A., Ruiter, A., Eds.; Reed Business: Amsterdam, The Netherlands, 2011; pp. 31–70. (In Dutch) [Google Scholar]
- Bergwerff, A.A.; Van Dam, G.J.; Rotmans, J.P.; Deelder, A.M.; Kamerling, J.P.; Vliegenthart, J.F. The immunologically reactive part of immunopurified circulating anodic antigen from Schistosoma mansoni is a threonine-linked polysaccharide consisting of --> 6)-(beta-D-GlcpA-(1 --> 3))-beta-D-GalpNAc-(1 --> repeating units. J. Biol. Chem. 1994, 269, 31510–31517. [Google Scholar] [CrossRef]
- Sun, G.G.; Wang, Z.Q.; Liu, C.Y.; Jiang, P.; Liu, P.-D.; Wen, H.; Qi, X.; Wang, L.; Cui, J. Early serodiagnosis of trichinellosis by ELISA using excretory–secretory antigens of Trichinella spiralis adult worms. Parasites Vectors 2015, 8, 484. [Google Scholar] [CrossRef] [Green Version]
- Morales-Yanez, F.J.; Sariego, I.; Vincke, C.; Hassanzadeh-Ghassabeh, G.; Polman, K.; Muyldermans, S. An innovative approach in the detection of Toxocara canis excretory/secretory antigens using specific nanobodies. Int. J. Parasitol. 2019, 49, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Roitt, I.; Brostoff, J.; Male, D. Immunology, 6th ed.; Mosby: London, UK, 2001; p. 480. [Google Scholar]
- Janeway, C.A., Jr.; Travers, P.; Walport, M.; Shlomchik, M.J. Immunobiology: The Immunesystem in Health and Disease, 6th ed.; Elsevier Ltd.: Oxford, UK, 2005; p. 823. [Google Scholar]
- Bruschi, F.; Gómez-Morales, M.A.; Hill, D.E. International Commission on Trichinellosis: Recommendations on the use of serological tests for the detection of Trichinella infection in animals and humans. Food Waterborne Parasitol. 2019, 14, e00032. [Google Scholar] [CrossRef]
- Leavy, O. The birth of monoclonal antibodies. Nat. Immunol. 2016, 17, S13. [Google Scholar] [CrossRef]
- Ehrlich, P.H.; Moyle, W.R. Cooperative immunoassays: Ultrasensitive assays with mixed monoclonal antibodies. Science 1983, 221, 279–281. [Google Scholar] [CrossRef]
- Marega, R.; Desroche, N.; Huet, A.-C.; Paulus, M.; Suarez Pantaleon, C.; Larose, D.; Arbault, P.; Delahaut, P.; Gillard, N. A general strategy to control antibody specificity against targets showing molecular and biological similarity: Salmonella case study. Sci. Rep. 2020, 10, 18439. [Google Scholar] [CrossRef] [PubMed]
- Petrenko, V.A.; Vodyanoy, V.J. Phage display for detection of biological threat agents. J. Microbiol. Methods 2003, 53, 253–262. [Google Scholar] [CrossRef]
- Nguyen, X.H.; Trinh, T.-L.; Vu, T.-B.-H.; Le, Q.-H.; To, K.-A. Isolation of phage-display library-derived scFv antibody specific to Listeria monocytogenes by a novel immobilized method. J. Appl. Microbiol. 2018, 124, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, R.F. The foundations of immunochemistry. In The Immunoassay Handbook—Theory and Applications of Ligand Binding, ELISA and Related Techniques, 4th ed.; Wild, D., Ed.; Elsevier: Amsterdam, The Netherlands, 2013; Chapter 4.1; pp. 339–356. [Google Scholar] [CrossRef]
- Swildens, B. Detection and Transmission of Extracellular Factor Producing Streptococcus suis Serotype 2 Strains in Pigs. Ph.D. Thesis, Utrecht University, Utrecht, The Netherlands, 2009. [Google Scholar]
- Brown, E.; Lawson, S.; Welbon, C.; Gnanandarajah, J.; Li, J.; Murtaugh, M.P.; Nelson, E.A.; Molina, R.M.; Zimmerman, J.J.; Rowland, R.R.R.; et al. Antibody response to porcine reproductive and respiratory syndrome virus (PRRSV) nonstructural proteins and implications for diagnostic detection and differentiation of PRRSV types I and II. Clin. Vaccine Immunol. 2009, 16, 628–635. [Google Scholar] [CrossRef] [Green Version]
- World Organization for Animal Health. Campylobacter jejuni and Campylobacter coli. In OIE Manual of Diagnostic Tests and Vaccines for Terrestrial Animals (Mammals, Birds and Bees), 6th ed.; OIE Biological Standards Commission, Ed.; Office International Des Epizooties: Paris, France, 2008; Volume 2, Chapter 2.9.3; pp. 1185–1191. [Google Scholar]
- AbuOun, M.; Manning, G.; Cawthraw, S.A.; Ridley, A.; Ahmed, I.H.; Wassenaar, T.M.; Newell, D.G. Cytolethal distending toxin (CDT)-negative Campylobacter jejuni strains and anti-CDT neutralizing antibodies are induced during human infection but not during colonization in chickens. Infect. Immun. 2005, 73, 3053–3062. [Google Scholar] [CrossRef] [Green Version]
- Berndt, A.; Wilhelm, A.; Jugert, C.; Pieper, J.; Sachse, K.; Methner, U. Chicken cecum immune response to Salmonella enterica serovars of different levels of invasiveness. Infect. Immun. 2007, 75, 5993–6007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berk, P.A.; Van der Heijden, H.M.J.F.; Mooijman, K.A. On Behalf of the European CommissionComparability of Different ELISAs on the Detection of Salmonella spp. Antibodies in Meat Juice and Serum, RIVM Report 330604007. 2008. Available online: https://www.rivm.nl/bibliotheek/rapporten/330604007.pdf (accessed on 14 December 2020).
- Vico, J.P.; Mainar-Jaime, R.C. The use of meat juice or blood serum for the diagnosis of Salmonella infection in pigs and its possible implications on Salmonella control programs. J. Vet. Diagn. Investig. 2011, 23, 528–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mousing, J.; Thode Jensen, P.; Halgaard, C.; Bager, F.; Feld, N.; Nielsen, B.; Nielsen, J.P.; Beth-Nielsen, S. Nation-wide Salmonella enterica surveillance and control in Danish slaughter swine herds. Prev. Vet. Med. 1997, 29, 247–261. [Google Scholar] [CrossRef]
- Meemken, D.; Tangemann, A.H.; Meermeier, D.; Gundlach, S.; Mischok, D.; Greiner, M.; Klein, G.; Blaha, T. Establishment of serological herd profiles for zoonoses and production diseases in pigs by “meat juice multi-serology”. Prev. Vet. Med. 2014, 113, 589–598. [Google Scholar] [CrossRef]
- Dzierzon, J.; Oswaldi, V.; Merle, R.; Langkabel, N.; Meemken, D. High Predictive Power of Meat Juice Serology on the Presence of Hepatitis E Virus in Slaughter Pigs. Foodborne Pathog. Dis. 2020, 17, 687–692. [Google Scholar] [CrossRef]
- Berger-Schoch, A.E.; Bernet, D.; Doherr, M.G.; Gottstein, B.; Frey, C.F. Toxoplasma gondii in Switzerland: A serosurvey based on meat juice analysis of slaughtered pigs, wild boar, sheep and cattle. Zoonoses Public Health 2011, 58, 472–478. [Google Scholar] [CrossRef] [PubMed]
- Gazzonis, A.L.; Zanzani, S.A.; Villa, L.; Manfredi, M.T. Toxoplasma gondii infection in meat-producing small ruminants: Meat juice serology and genotyping. Parasitol. Int. 2020, 76, 102060. [Google Scholar] [CrossRef] [Green Version]
- Wallander, C.; Frössling, J.; Vågsholm, I.; Burrells, A.; Lundén, A. “Meat juice” is not a homogeneous serological matrix. Foodborne Pathog. Dis. 2015, 12, 280–288. [Google Scholar] [CrossRef]
- Hill, D.E.; Chirukandoth, S.; Dubey, J.P.; Lunney, J.K.; Gamble, H.R. Comparison of detection methods for Toxoplasma gondii in naturally and experimentally infected swine. Vet. Parasitol. 2006, 141, 9–17. [Google Scholar] [CrossRef]
- Atkinson, B.M.; Bearson, B.L.; Loving, C.L.; Zimmerman, J.J.; Kich, J.D.; Bearson, S.M.D. Detection of Salmonella-specific antibody in swine oral fluids. Porc. Health Manag. 2019, 5, 29. [Google Scholar] [CrossRef]
- Prickett, J.R.; Zimmerman, J.J. The development of oral fluid-based diagnostics and applications in veterinary medicine. Anim. Health Res. Rev. 2010, 11, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Bjustrom-Kraft, J.; Christopher-Hennings, J.; Daly, R.; Main, R.; Torrison, J.; Thurn, M.; Zimmerman, J. The use of oral fluid diagnostics in swine medicine. J. Swine Health Prod. 2018, 26, 262–269. [Google Scholar]
- Fosgate, G.T. Practical sample size calculations for surveillance and diagnostic investigations. J. Vet. Diagn. Investig. 2009, 21, 3–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freuling, C.M.; Müller, T.F.; Mettenleiter, T.C. Vaccines against pseudorabies virus (PrV). Vet. Microbiol. 2017, 206, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Leifer, I.; Everett, H.; Hoffmann, B.; Sosan, O.; Crooke, H.; Beer, M.; Blome, S. Escape of classical swine fever C-strain vaccine virus from detection by C-strain specific real-time RT-PCR caused by a point mutation in the primer-binding site. J. Virol. Methods 2010, 166, 98–100. [Google Scholar] [CrossRef]
- Silva, N.F.D.; Neves, M.M.P.S.; Magalhães, J.M.C.S.; Freire, C.; Delerue-Matos, C. Electrochemical immunosensor towards invasion-associated protein p60: An alternative strategy for Listeria monocytogenes screening in food. Talanta 2020, 120976. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, S.; Amamcharla, J.; Shin, J.E. Possible application of electronic nose systems for meat safety: An overview. In Electronic Noses and Tongues in Food Science, 1st ed.; Rodriguez, M.M.L., Ed.; Part I; Academic Press: London, UK, 2016; Chapter 7; pp. 59–71. [Google Scholar]
- Gorski, L. Selective enrichment media bias the types of Salmonella enterica strains isolated from mixed strain cultures and complex enrichment broths. PLoS ONE 2012, 7, e34722. [Google Scholar] [CrossRef]
- Löfström, C.; Krause, M.; Josefsen, M.H.; Hansen, F.; Hoorfar, J. Validation of a same-day real-time PCR method for screening of meat and carcass swabs for Salmonella. BMC Microbiol. 2009, 9, 85. [Google Scholar] [CrossRef] [Green Version]
- Delibato, E.; Rodriguez-Lazaro, D.; Gianfranceschi, M.; De Cesare, A.; Comin, D.; Gattuso, A.; Hernandez, M.; Sonnessa, M.; Pasquali, F.; Sreter-Lancz, Z.; et al. European validation of Real-Time PCR method for detection of Salmonella spp. in pork meat. Int. J. Food Microbiol. 2014, 184, 134–138. [Google Scholar] [CrossRef]
- Srisa-Art, M.; Boehle, K.E.; Geiss, B.J.; Henry, C.S. Highly sensitive detection of Salmonella typhimurium using a colorimetric paper-based analytical device coupled with immunomagnetic separation. Anal. Chem. 2018, 90, 1035–1043. [Google Scholar] [CrossRef]
- Brewster, J.D. Isolation and concentration of Salmonellae with an immunoaffinity column. J. Microbiol. Methods 2003, 55, 287–293. [Google Scholar] [CrossRef]
- Lee, K.-M.; Runyon, M.; Herrman, T.J.; Phillips, R.; Hsieh, J. Review of Salmonella detection and identification methods: Aspects of rapid emergency response and food safety. Food Control 2015, 47, 264–276. [Google Scholar] [CrossRef]
- Saengthongpinit, C.; Bergwerff, A.A.; Van Knapen, F.; Amavisit, P.; Sirinarumitr, T.; Sakpuaram, T. Comparison of immunomagnetic separation and multiplex PCR assay for detection of Campylobacter jejuni and Campylobacter coli in chicken meat. Kasetsart J. 2007, 41, 696–704. [Google Scholar]
- Ozalp, V.C.; Bayramoglu, G.; Erdem, Z.; Arica, M.Y. Pathogen detection in complex samples by quartz crystal microbalance sensor coupled to aptamer functionalized core–shell type magnetic separation. Anal. Chim. Acta 2015, 853, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Huo, X.; Chen, Q.; Wang, L.; Cai, G.; Qi, W.; Xia, Z.; Wen, W.; Lin, J. Continuous-flow separation and efficient concentration of foodborne bacteria from large volume using nickel nanowire bridge in microfluidic chip. Micromachines 2019, 10, 644. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-C.; Tu, Y.-Y.; Chang, H.-M. Thermal stability of bovine milk immunoglobulin g (igg) and the effect of added thermal protectants on the stability. J. Food Sci. 2000, 65, 188–193. [Google Scholar] [CrossRef]
- Bergwerff, A.A. New strategies for detecting Salmonella and sulphonamides. World Poult. 2006, 22, 2–3. [Google Scholar]
- Videnska, P.; Sisak, F.; Havlickova, H.; Faldynova, M.; Rychlik, I. Influence of Salmonella enterica serovar Enteritidis infection on the composition of chicken cecal microbiota. BMC Vet. Res. 2013, 9, 140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blondel, C.J.; Yang, H.-J.; Castro, B.; Chiang, S.; Toro, C.S.; Zaldívar, M.; Contreras, I.; Andrews-Polymenis, H.L.; Santiviago, C.A. Contribution of the Type VI Secretion System Encoded in SPI-19 to Chicken Colonization by Salmonella enterica Serotypes Gallinarum and Enteritidis. PLoS ONE 2010, 5, e11724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kothary, M.H.; Babu, U.S. Infective dose of foodborne pathogens in volunteers: A review. J. Food Saf. 2001, 21, 49–73. [Google Scholar] [CrossRef]
- Smith, J.L. Infectious dose and an aging population: Susceptibility of the aged to foodborne pathogens. In Foodborne Pathogens, Food Microbiology and Food Safety; Gurtler, J., Doyle, M., Kornacki, J., Eds.; Springer: Cham, Switzerland, 2017; pp. 451–468. [Google Scholar] [CrossRef]
- Todd, E.C.D.; Greig, J.D.; Bartleson, C.A.; Michaels, B.S. Outbreaks where food workers have been implicated in the spread of foodborne disease. Part 4. Infective doses and pathogen carriage. J. Food Prot. 2008, 71, 2339–2373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mead, G.; Lammerding, A.M.; Cox, N.; Doyle, M.P.; Humbert, F.; Kulikovskiy, A.; Panin, A.; Pinheiro Do Nascimento, V.; Wierup, M. The Salmonella on Raw Poultry Writing Committee. Scientific and technical factors affecting the setting of Salmonella criteria for raw poultry: A global perspective. J. Food Prot. 2010, 73, 1566–1590. [Google Scholar] [CrossRef] [Green Version]
- Eggenkamp, A.E.; Bergwerff, A.A. Microbiologie in het laboratorium. In Inleiding tot de Levensmiddelenhygiëne—Achtergrond en Feiten, 2nd ed.; Lipman, L.J.A., Ruiter, A., Eds.; Reed Business: Amsterdam, The Netherlands, 2011; pp. 144–165. (In Dutch) [Google Scholar]
- Reist, M.; Jemmi, T.; Stärk, K.D.C. Policy-driven development of cost-effective, risk-based surveillance strategies. Prev. Vet. Med. 2012, 105, 176–184. [Google Scholar] [CrossRef]
- Martin, S.W.; Shoukri, M.; Thorburn, M.A. Evaluating the health status of herds based on tests applied to individuals. Prev. Vet. Med. 1992, 14, 33–43. [Google Scholar] [CrossRef]
- Sahin, O.; Kassem, I.I.; Shen, Z.; Lin, J.; Rajashekara, G.; Zhang, Q. Campylobacter in poultry: Ecology and potential interventions. Avian Dis. 2015, 59, 185–200. [Google Scholar] [CrossRef]
- Hugas, M.; Tsigarida, E. Pros and cons of carcass decontamination: The role of the European Food Safety Authority. Meat Sci. 2008, 78, 43–52. [Google Scholar] [CrossRef]
- European Food Safety Authority Panel on Biological Hazards; Ricci, A.; Allende, A.; Bolton, D.; Chemaly, M.; Davies, R.; Fernández Escámez, P.S.; Girones, R.; Herman, L.; Koutsoumanis, K.; et al. Scientific opinion on the guidance on the requirements for the development of microbiological criteria. EFSA J. 2017, 15, 5052. [Google Scholar] [CrossRef] [Green Version]
- Barlow, S.M.; Boobis, A.R.; Bridges, J.; Cockburn, A.; Dekant, W.; Hepburn, P.; Houben, G.F.; König, J.; Nauta, M.J.; Schuermans, J.; et al. The role of hazard- and risk-based approaches in ensuring food safety. Trends Food Sci. Technol. 2015, 46, 176–188. [Google Scholar] [CrossRef] [Green Version]
- Bahrndorff, S.; Rangstrup-Christensen, L.; Nordentoft, S.; Hald, B. Foodborne Disease Prevention and Broiler Chickens with Reduced Campylobacter Infection. Emerg. Infect. Dis. 2013, 19, 425–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finley, R.; Reid-Smith, R.; Weese, J.S. Human health implications of Salmonella-contaminated natural pet treats and raw pet food. Clin. Infect. Dis. 2006, 42, 686–691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Notermans, S.; Wernars, K. Immunological methods for detection of foodborne pathogens and their toxins. Int. J. Food Microbiol. 1991, 12, 91–102. [Google Scholar] [CrossRef]
- Candlish, A.A.G. Immunological methods in food microbiology. Food Microbiol. 1991, 8, 1–14. [Google Scholar] [CrossRef]
- Wild, D. (Ed.) The Immunoassay Handbook, 4th ed.; Elsevier: Amsterdam, The Netherlands, 2013. [Google Scholar] [CrossRef]
- Köhler, W. Zentralblatt fur Bakteriologie—100 years ago Agglutination and filament formation of Proteus bacteria and maternal-fetal transfer of agglutinins. Int. J. Med. Microbiol. 2001, 290, 643–646. [Google Scholar] [CrossRef]
- Gracias, K.S.; McKillip, J.L. A review of conventional detection and enumeration methods for pathogenic bacteria in food. Can. J. Microbiol. 2004, 50, 883–890. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, B.; Sankaran, S.; Stegemann, L.; Strassert, C.A.; Jonkheijm, P.; Voskuhl, J. Agglutination of bacteria using polyvalent nanoparticles of aggregation-induced emissive thiophthalonitrile dyes. J. Mater. Chem. B 2016, 4, 4732–4738. [Google Scholar] [CrossRef] [Green Version]
- Denayer, S.; Delbrassinne, L.; Nia, Y.; Botteldoorn, N. Food-borne outbreak investigation and molecular typing: High diversity of Staphylococcus aureus strains and importance of toxin detection. Toxins 2017, 9, 407. [Google Scholar] [CrossRef] [Green Version]
- Abel, E.S. Preliminary studies on the detection of Bacillus cereus and its toxins: Comparing conventional and immunological assays with a direct polymerase chain reaction method. Curr. J. Appl. Sci. Technol. 2019, 36, 1–9. [Google Scholar] [CrossRef]
- Fletcher, P.; Logan, N.A. Improved cytotoxicity assay for Bacillus cereus diarrhoeal enterotoxin. Lett. Appl. Microbiol. 1999, 28, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Chart, H.; Willshaw, G.A.; Cheasty, T. Evaluation of a reversed passive latex agglutination test for the detection of verocytotoxin (VT) expressed by strains of VT-producing Escherichia coli. Lett. Appl. Microbiol. 2001, 32, 370–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Farrell, B. Evolution in lateral flow-based immunoassay systems. In Lateral Flow Immunoassay; Wong, R., Tse, H., Eds.; Springer Science & Business Media: New York, NY, USA, 2008; pp. 1–34. [Google Scholar]
- Bird, C.B.; Hoerner, R.J.; Restaino, L. Reveal 8–Hour test system for detection of Escherichia coli O157:H7 in raw ground beef, raw beef cubes, and lettuce rinse: Collaborative study. J. AOAC Int. 2001, 84, 719–736. [Google Scholar] [CrossRef] [Green Version]
- Bird, C.B.; Hoerner, R.J.; Restaino, L. Comparison of the Reveal 20–hour method and the BAM culture method for the detection of Escherichia coli O157:H7 in selected foods and environmental swabs: Collaborative study. J. AOAC Int. 2001, 84, 737–751. [Google Scholar] [CrossRef] [Green Version]
- Feldsine, P.T.; Falbo-Nelson, M.T.; Brunelle, S.L.; Forgey, R.L. Visual Immunoprecipitate Assay (VIP) for Detection of Enterohemorrhagic Escherichia coli (EHEC) 0157:H7 in Selected Foods: Collaborative Study. J. AOAC Int. 1997, 80, 517–529. [Google Scholar] [CrossRef] [Green Version]
- Feldsine, P.T.; Mui, L.A.; Forgey, R.L.; Kerr, D.E.A. Equivalence of Visual Immunoprecipitate Assay (VIP®) for Salmonella for the Detection of Motile and Nonmotile Salmonella in All Foods to AOAC Culture Method: Collaborative Study. J. AOAC Int. 2000, 83, 888–902. [Google Scholar] [CrossRef] [Green Version]
- Feldsine, P.T.; Kerr, D.E.; Shen, G.; Lienau, A. Comparative Validation Study to Demonstrate the Equivalence of a Minor Modification to AOAC Method 997.03 Visual Immunoprecipitate (VIP®) for Listeria to the Reference Culture Method. J. AOAC Int. 2009, 92, 1421–1425. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Macdonald, J. Multiplexed lateral flow biosensors: Technological advances for radically improving point-of-care diagnoses. Biosens. Bioelectron. 2016, 83, 177–192. [Google Scholar] [CrossRef]
- Chang, L.; Li, J.; Wang, L. Immuno-PCR: An ultrasensitive immunoassay for biomolecular detection. Anal. Chim. Acta 2016, 910, 12–24. [Google Scholar] [CrossRef]
- Sadat Tabatabaei, M.; Islam, R.; Ahmed, M. Applications of gold nanoparticles in ELISA, PCR, and immuno-PCR assays: A review. Anal. Chim. Acta 2021, 1143, 250–266. [Google Scholar] [CrossRef]
- Rajkovic, A.; El Moualij, B.; Fikri, Y.; Dierick, K.; Zorzi, W.; Heinen, E.; Uner, A.; Uyttendaele, M. Detection of Clostridium botulinum neurotoxins A and B in milk by ELISA and immuno-PCR at higher sensitivity than mouse bio-assay. Food Anal. Methods 2012, 5, 319–326. [Google Scholar] [CrossRef]
- Rajkovic, A.; El Moualij, B.; Uyttendaele, M.; Brolet, P.; Zorzi, W.; Heinen, E.; Foubert, E.; Debevere, J. Immunoquantitative Real-Time PCR for Detection and Quantification of Staphylococcus aureus Enterotoxin B in Foods. Appl. Environ. Microbiol. 2006, 72, 6593–6599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, A.; Von Eiff, C.; Kuczius, T.; Omoe, K.; Peters, G.; Becker, K. A quantitative real-time immuno-PCR approach for detection of staphylococcal enterotoxins. J. Mol. Med. 2007, 85, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Grangar, V.; Curiale, M.S.; D’onorio, A.; Schultz, A.; Johnson, R.L.; Atrache, V. VIDAS® enzyme-linked Immunofluorescent assay for detection of Listeria in Foods: Collaborative study. J. AOAC Int. 2000, 83, 903–918. [Google Scholar] [CrossRef] [Green Version]
- Karuppannan, A.K.; De Castro, A.M.M.G.; Opriessnig, T. Recent advances in veterinary diagnostic virology. In Advanced Techniques in Diagnostic Microbiology, 3rd ed.; Tang, Y.W., Stratton, C., Eds.; Applications; Springer: Cham, Switzerland, 2018; Volume 2, pp. 317–344. [Google Scholar] [CrossRef]
- Olopoenia, L.A.; King, A.L. Widal agglutination test—100 years later: Still plagued by controversy. Postgrad Med. J. 2000, 76, 80–84. [Google Scholar] [CrossRef]
- Kennedy, D.; Wilkinson, M.G. Application of flow cytometry to the detection of pathogenic bacteria. Curr. Issues Mol. Biol. 2017, 23, 21–38. [Google Scholar] [CrossRef] [Green Version]
- Robinson, J.P. Overview of flow cytometry and microbiology. Curr. Protoc. Cytom. 2018, 84, e37. [Google Scholar] [CrossRef]
- Reslova, N.; Michna, V.; Kasny, M.; Mikel, P.; Kralik, P. xMAP technology: Applications in detection of pathogens. Front. Microbiol. 2017, 8, 55. [Google Scholar] [CrossRef]
- Vignali, D.A.A. Multiplexed particle-based flow cytometric assays. J. Immunol. Methods 2000, 243, 243–255. [Google Scholar] [CrossRef]
- Kellar, K.L.; Iannone, M.A. Multiplexed microsphere-based flow cytometric assays. Exp. Hematol. 2002, 30, 1227–1237. [Google Scholar] [CrossRef]
- Graham, H.; Chandler, D.J.; Dunbar, S.A. The genesis and evolution of bead-based multiplexing. Methods 2019, 158, 2–11. [Google Scholar] [CrossRef]
- Foroutan Parsa, S.; Vafajoo, A.; Rostami, A.; Salarian, R.; Rabiee, M.; Rabiee, N.; Rabiee, G.; Tahriri, M.; Yadegari, A.; Vashaee, D.; et al. Early diagnosis of disease using microbead array technology: A review. Anal. Chim. Acta 2018, 1032, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Christopher-Hennings, J.; Araujo, K.P.C.; Souza, C.J.H.; Fang, Y.; Lawson, S.; Nelson, E.A.; Clement, T.; Dunn, M.; Lunney, J.K. Opportunities for bead-based multiplex assays in veterinary diagnostic laboratories. J. Vet. Diagn. Investig. 2013, 25, 671–691. [Google Scholar] [CrossRef] [Green Version]
- Shepelyakovskaya, A.; Rudenko, N.; Karatovskaya, A.; Shchannikova, M.; Shulcheva, I.; Fursova, K.; Zamyatina, A.; Boziev, K.; Oleinikov, V.; Brovko, F. Development of a bead-based multiplex assay for the simultaneous quantification of three staphylococcal enterotoxins in food by flow cytometry. Food Anal. Methods 2020, 13, 1202–1210. [Google Scholar] [CrossRef]
- Mechaly, A.; Vitner, E.; Levy, H.; Weiss, S.; Bar-David, E.; Gur, D.; Koren, M.; Cohen, H.; Cohen, O.; Mamroud, E.; et al. Simultaneous immunodetection of anthrax, plague, and tularemia from blood cultures by use of multiplexed suspension arrays. J. Clin. Microbiol. 2018, 56, e01479-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leach, K.M.; Stroot, J.M.; Lim, D.V. Same-day detection of Escherichia coli O157:H7 from spinach by using electrochemiluminescent and cytometric bead array biosensors. Appl. Environ. Microbiol. 2010, 76, 8044–8052. [Google Scholar] [CrossRef] [Green Version]
- Liebsch, C.; Rödiger, S.; Böhm, A.; Nitschke, J.; Weinreich, J.; Fruth, A.; Roggenbuck, D.; Lehmann, W.; Schedler, U.; Juretzek, T.; et al. Solid-phase microbead array for multiplex O-serotyping of Escherichia coli. Microchim. Acta 2017, 184, 1405–1415. [Google Scholar] [CrossRef]
- Bergwerff, A.A.; Bokken, G.C.A.M.; Gortemaker, B.G.M. Immobilisation of Antigenic Carbohydrates to Support Detection of Pathogenic Microorganisms. Patent Application U.S. 20090081638 A1, 26 March 2009. Available online: https://patents.google.com/patent/US20090081638A1/en#patentCitations (accessed on 18 January 2021).
- Van der Wal, F.J.; Achterberg, R.P.; Maassen, C.B.M. A bead-based suspension array for the detection of Salmonella antibodies in pig sera. BMC Vet. Res. 2018, 14, 226. [Google Scholar] [CrossRef] [Green Version]
- Thomas, M.E.; Klinkenberg, D.; Bergwerff, A.A.; Van Eerden, E.; Stegeman, J.A.; Bouma, A. Evaluation of suspension array analysis for detection of egg yolk antibodies against Salmonella Enteritidis. Prev. Vet. Med. 2010, 95, 137–143. [Google Scholar] [CrossRef]
- European Commission of the European Union. Regulation (EU) 2015/1375 of 10 August 2015 laying down specific rules on official controls for Trichinella in meat. Off. J. Eur. Union 2015, L212, 7. [Google Scholar]
- Bokken, G.C.A.M. Concurrent Monitoring of Trichinella and Toxoplasma Infections in Pigs from Controlled Housing Systems. Ph.D. Thesis, Utrecht University, Utrecht, The Netherlands, 2017. Available online: https://dspace.library.uu.nl/handle/1874/351669 (accessed on 15 January 2021).
- Jiang, P.; Wang, Z.-Q.; Cui, J.; Zhang, X. Comparison of artificial digestion and Baermann’s methods for detection of Trichinella spiralis pre-encapsulated larvae in muscles with low-level infections. Foodborne Pathog. Dis. 2012, 9, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Bokken, G.C.A.M.; Bergwerff, A.A.; Van Knapen, F. A novel bead-based assay to detect specific antibody responses against Toxoplasma gondi and Trichinella spiralis simultaneously in sera of experimentally infected swine. BMC Vet. Res. 2012, 8, 36. [Google Scholar] [CrossRef] [Green Version]
- Havelaar, A.H.; Van Rosse, F.; Bucura, C.; Toetenel, M.A.; Haagsma, J.A.; Kurowicka, D.; Heesterbeek, J.A.P.; Speybroeck, N.; Langelaar, M.F.M.; Van der Giessen, J.W.B.; et al. Prioritizing emerging zoonoses in the Netherlands. PLoS ONE 2010, 5, e13965. [Google Scholar] [CrossRef]
- Batz, M.; Hoffmann, S.; Morris, J., Jr. Ranking the Risks: The 10 Pathogen-Food Combinations with the Greatest Burden on Public Health, Emerging Pathogens Institute at University of Florida. 2011. Available online: http://hdl.handle.net/10244/1022 (accessed on 15 January 2021).
- Guilbault, G.; Kramer, D.; Cannon, P., Jr. Electrical determination of organophosphorous compounds. Anal. Chem. 1962, 34, 1437–1439. [Google Scholar] [CrossRef]
- Duffy, G.F.; Moore, E.J. Electrochemical immunosensors for food analysis: A review of recent developments. Anal. Lett. 2017, 50, 1–32. [Google Scholar] [CrossRef]
- Evtugyn, G.A.; Shamagsumova, R.V.; Hianik, T. Biosensors for detection mycotoxins and pathogenic bacteria in food. In Nanobiosensors; Grumezescu, A.M., Ed.; Academic Press: London, UK, 2017; pp. 35–92. [Google Scholar] [CrossRef]
- Cinti, S.; Volpe, G.; Piermarini, S.; Delibato, E.; Palleschi, G. Electrochemical biosensors for rapid detection of foodborne Salmonella: A critical overview. Sensors 2017, 17, 1910. [Google Scholar] [CrossRef]
- Bhardwaj, N.; Bhardwaj, S.K.; Nayak, M.K.; Mehta, J.; Kim, K.-H.; Deep, A. Fluorescent nanobiosensors for the targeted detection of foodborne bacteria. Trends Anal. Chem. 2017, 97, 120e135. [Google Scholar] [CrossRef]
- Das, B.; Balasubramanian, P.; Jayabalan, R.; Lekshmi, N.; Thomas, S. Strategies behind biosensors for food and waterborne pathogens. In Quorum Sensing and Its Biotechnological Applications; Kalia, V., Ed.; Springer: Singapore, 2018; pp. 107–141. [Google Scholar] [CrossRef]
- Ali, A.A.; Altemimi, A.B.; Alhelfi, N.; Ibrahim, S.A. Application of biosensors for detection of pathogenic food bacteria: A review. Biosensors 2020, 10, 58. [Google Scholar] [CrossRef]
- Yunus, G.; Kuddus, M. Electrochemical biosensor for food borne pathogens: An overview. Carpath. J. Food Sci. Technol. 2020, 12, 5–16. [Google Scholar] [CrossRef]
- Silva, N.F.D.; Magalhães, J.M.C.S.; Freire, C.; Delerue-Matos, C. Electrochemical biosensors for Salmonella: State of the art and challenges in food safety assessment. Biosens. Bioelectron. 2018, 99, 667–682. [Google Scholar] [CrossRef] [Green Version]
- Riu, J.; Giussani, B. Electrochemical biosensors for the detection of pathogenic bacteria in food. Trends Anal. Chem. 2020, 126, 115863. [Google Scholar] [CrossRef]
- Liébana, A.; Brandão, D.; Alegret, S.; Pividori, M.I. Electrochemical immunosensors, genosensors and phagosensors for Salmonella detection. Anal. Methods 2014, 6, 8858–8873. [Google Scholar] [CrossRef]
- Khansili, N.; Rattu, G.; Krishna, P.M. Label-free optical biosensors for food and biological sensor applications. Sens. Actuators B Chem. 2018, 265, 35–49. [Google Scholar] [CrossRef]
- Zourob, M.; Elwary, S.; Turner, A. (Eds.) Principles of Bacterial Detection: Biosensors, Recognition Receptors and Microsystems; Springer: New York, NY, USA, 2008. [Google Scholar] [CrossRef]
- Gehring, A.G.; Tu, S.-I. High-throughput biosensors for multiplexed food-borne pathogen detection. Annu. Rev. Anal. Chem. 2011, 4, 151–172. [Google Scholar] [CrossRef] [PubMed]
- Bozal-Palabiyik, B.; Gumustas, A.; Ozkan, S.A.; Uslu, B. Biosensor-based methods for the determination of foodborne pathogens. In Handbook of Food Bioengineering—Foodborne Diseases; Holban, A.M., Grumezescu, A.M., Eds.; Academic Press: London, UK, 2018; Chapter 12; pp. 379–420. [Google Scholar] [CrossRef]
- Kumar Mishra, G.; Barfidokht, A.; Tehrani, F.; Kumar Mishra, R. Food Safety Analysis Using Electrochemical Biosensors. Foods 2018, 7, 141. [Google Scholar] [CrossRef] [Green Version]
- Poltronieri, P.; Mezzolla, V.; Primiceri, E.; Maruccio, G. Biosensors for the detection of food pathogens. Foods 2014, 3, 511–526. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information (PubMed). Available online: https://pubmed.ncbi.nlm.nih.gov (accessed on 20 November 2020).
- Bergwerff, A.A. Moderne microbiologische technieken. In Inleiding tot de—Achtergrond en Feiten, 2nd ed.; Lipman, L.J.A., Ruiter, A., Eds.; Reed Business: Amsterdam, The Netherlands, 2011; pp. 167–178. (In Dutch) [Google Scholar]
- Situ, C.; Mooney, M.H.; Elliott, C.T.; Buijs, J. Advances in surface plasmon resonance biosensor technology towards high-throughput, food-safety analysis. Trends Anal. Chem. 2010, 29, 1305–1315. [Google Scholar] [CrossRef]
- Bokken, G.C.A.M.; Corbee, R.J.; Van Knapen, F.; Bergwerff, A.A. Immunochemical detection of Salmonella group B, D and E using an optical surface plasmon resonance biosensor. FEMS Microbiol. Lett. 2003, 222, 75–82. [Google Scholar] [CrossRef] [Green Version]
- Taylor, A.D.; Ladd, J.; Homola, J.; Jiang, S. Surface plasmon resonance (SPR) sensors for the detection of bacterial pathogens. In Principles of Bacterial Detection: Biosensors, Recognition Receptors and Microsystems; Zourob, M., Elwary, S., Turner, A., Eds.; Springer: New York, NY, USA, 2008; Part II; Chapter 5; pp. 83–108. [Google Scholar] [CrossRef]
- Bergwerff, A.A.; Van Knapen, F. Surface plasmon resonance biosensors for detection of pathogenic microorganisms: Strategies to secure food and environmental safety. J. AOAC Int. 2006, 89, 826–831. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Knoll, W.; Dostalek, J. Bacterial pathogen surface plasmon resonance biosensor advanced by long range surface plasmons and magnetic nanoparticle assays. Anal. Chem. 2012, 84, 8345–8350. [Google Scholar] [CrossRef] [PubMed]
- Jongerius-Gortemaker, B.G.M.; Goverde, R.L.J.; Van Knapen, F.; Bergwerff, A.A. Surface plasmon resonance (BIACORE) detection of serum antibodies against Salmonella enteritidis and Salmonella typhimurium. J. Immunol. Meth. 2002, 266, 33–44. [Google Scholar] [CrossRef]
- Thomas, A.; Bouma, A.; Van Eerden, E.; Landman, W.J.M.; Van Knapen, F.; Stegeman, A.; Bergwerff, A.A. Detection of egg yolk antibodies reflecting Salmonella enteritidis infections using a surface plasmon resonance biosensor. J. Immunol. Methods 2006, 315, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Datta Mazumdar, S.; Barlen, B.; Kramer, T.; Keusgen, M. A rapid serological assay for prediction of Salmonella infection status in slaughter pigs using surface plasmon resonance. J. Microbiol. Methods 2008, 75, 545–550. [Google Scholar] [CrossRef] [PubMed]
- Palchetti, I.; Mascini, M. Amperometric biosensors for pathogenic bacteria detection. In Principles of Bacterial Detection: Biosensors, Recognition Receptors and Microsystems; Zourob, M., Elwary, S., Turner, A., Eds.; Springer: New York, NY, USA, 2008; Part II; Chapter 13; pp. 299–312. [Google Scholar] [CrossRef]
- Melo, A.M.A.; Alexandre, D.L.; Furtado, R.F.; Borges, M.F.; Figueiredo, E.A.T.; Biswas, A.; Cheng, H.N.; Alves, C.R. Electrochemical immunosensors for Salmonella detection in food. Appl. Microbiol. Biotechnol. 2016, 100, 5301–5312. [Google Scholar] [CrossRef]
- Eissa, S.; Zourob, M. Ultrasensitive peptide-based multiplexed electrochemical biosensor for the simultaneous detection of Listeria monocytogenes and Staphylococcus aureus. Microchim. Acta 2020, 187, 486. [Google Scholar] [CrossRef] [PubMed]
- Wu, W. Inorganic nanomaterials for printed electronics: A review. Nanoscale 2017, 9, 7342–7372. [Google Scholar] [CrossRef]
- Viswanathan, S.; Ranib, C.; Ho, J.A. Electrochemical immunosensor for multiplexed detection of food-borne pathogens using nanocrystal bioconjugates and MWCNT screen-printed electrode. Talanta 2012, 94, 315–319. [Google Scholar] [CrossRef] [Green Version]
- Mohamed, H.M. Screen-printed disposable electrodes: Pharmaceutical applications and recent developments. Trends Anal. Chem. 2016, 82, 1–11. [Google Scholar] [CrossRef]
- Freitas, M.; Viswanathan, S.; Nouws, H.P.A.; Oliveira, M.B.P.P.; Delerue-Matos, C. Iron oxide/gold core/shell nanomagnetic probes and CdS biolabels for amplified electrochemical immunosensing of Salmonella typhimurium. Biosens. Bioelectron. 2014, 51, 195–200. [Google Scholar] [CrossRef] [Green Version]
- Birnbaumer, G.M.; Lieberzeit, P.A.; Richter, L.; Schirhagl, R.; Milnera, M.; Dickert, F.L.; Bailey, A.; Ertl, P. Detection of viruses with molecularly imprinted polymers integrated on a microfluidic biochip using contact-less dielectric microsensors. Lab. Chip 2009, 9, 3549–3556. [Google Scholar] [CrossRef] [PubMed]
- BelBruno, J.J. Molecularly imprinted polymers. Chem. Rev. 2019, 119, 94–119. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Zhou, Z.; Zhou, H.S. Molecularly imprinted polymers and surface imprinted polymers based electrochemical biosensor for infectious diseases. Sensors 2020, 20, 996. [Google Scholar] [CrossRef] [Green Version]
- Mukama, O.; Sinumvayo, J.P.; Shamoon, M.; Shoaib, M.; Mushimiyimana, H.; Safdar, W.; Bemena, L.; Rwibasira, P.; Mugisha, S.; Wang, Z. An update on aptamer-based multiplex system approaches for the detection of common foodborne pathogens. Food Anal. Methods 2017, 10, 2549–2565. [Google Scholar] [CrossRef]
- Majdinasab, M.; Hayat, A.; Marty, J.L. Aptamer-based assays and aptasensors for detection of pathogenic bacteria in food samples. Trends Anal. Chem. 2018, 107, 60–77. [Google Scholar] [CrossRef]
- Zhu, H.; Sikora, U.; Ozcan, A. Quantum dot enabled detection of Escherichia coli using a cell-phone. Analyst 2012, 137, 2541–2544. [Google Scholar] [CrossRef] [Green Version]
- Zeinhom, M.M.A.; Wang, Y.; Sheng, L.; Du, D.; Li, L.; Zhu, M.-J.; Lin, Y. Smart phone based immunosensor coupled with nanoflower signal amplification for rapid detection of Salmonella Enteritidis in milk, cheese and water. Sens. Actuators B Chem. 2018, 261, 75–82. [Google Scholar] [CrossRef]
- de Dieu Habimana, J.; Ji, J.; Sun, X. Minireview: Trends in optical-based biosensors for point-of-care bacterial pathogen detection for food safety and clinical diagnostics. Anal. Lett. 2018, 51, 2933–2966. [Google Scholar] [CrossRef]
- Kant, K.; Shahbazi, M.-A.; Dave, V.P.; Ngo, T.A.; Chidambara, V.A.; Than, L.Q.; Bang, D.D.; Wolff, A. Microfluidic devices for sample preparation and rapid detection of foodborne pathogens. Biotechnol. Adv. 2018, 36, 1003–1024. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, S.; Morarka, A.; Bodas, D.; Paknikar, K.M. Multiplexed detection of waterborne pathogens in circular microfluidics. Appl. Biochem. Biotechnol. 2012, 167, 1668–1677. [Google Scholar] [CrossRef]
- Dincer, C.; Bruch, R.; Kling, A.; Dittrich, P.S.; Urban, G.A. Multiplexed point-of-care testing—xPOCT. Trends Biotechnol. 2017, 35, 728–742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chin, C.; Laksanasopin, T.; Cheung, Y.; Steinmiller, D.; Linder, V.; Parsa, H.; Wang, J.; Moore, H.; Rouse, R.; Umviligihozo, G.; et al. Microfluidics-based diagnostics of infectious diseases in the developing world. Nat. Med. 2011, 17, 1015–1019. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, C.M.; Kinahan, D.J.; Mishra, R.; Mangwanya, F.; Kilcawley, N.; Ferreira, M.; Ducrée, J. Label-free, spatially multiplexed SPR detection of immunoassays on a highly integrated centrifugal Lab-on-a-Disc platform. Biosens. Bioelectron. 2018, 119, 86–93. [Google Scholar] [CrossRef]
- Posthuma-Trumpie, G.A.; Korf, J.; Van Amerongen, A. Lateral flow (immuno)assay: Its strengths, weaknesses, opportunities and threats. A literature survey. Anal. Bioanal. Chem. 2009, 393, 569–582. [Google Scholar] [CrossRef] [Green Version]
- Ahovan, Z.A.; Hashemi, A.; De Plano, L.M.; Gholipourmalekabadi, M.; Seifalian, A. Bacteriophage based biosensors: Trends, outcomes and challenges. Nanomaterials 2020, 10, 501. [Google Scholar] [CrossRef] [Green Version]
- Schofield, D.; Sharp, N.J.; Westwater, C. Phage-based platforms for the clinical detection of human bacterial pathogens. Bacteriophage 2012, 2, 105–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmelcher, M.; Loessner, M.J. Bacteriophage endolysins: Applications for food safety. Curr. Opin. Biotechnol. 2016, 37, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Zhao, X. Isothermal amplification technologies for the detection of foodborne pathogens. Food Anal. Methods 2018, 11, 1543–1560. [Google Scholar] [CrossRef]
- Garcia, J.L.; Gennari, S.M.; Machado, R.M.; Navarro, I.T. Toxoplasma gondii: Detection by mouse bioassay, histopathology, and polymerase chain reaction in tissues from experimentally infected pigs. Exp. Parasitol. 2006, 113, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Bezerra, R.A.; Carvalho, F.S.; Guimarães, L.A.; Rocha, D.S.; Silva, F.L.; Wenceslau, A.A.; Albuquerque, G.R. Comparison of methods for detection of Toxoplasma gondii in tissues of naturally exposed pigs. Parasitol. Res. 2012, 110, 509–514. [Google Scholar] [CrossRef]
- Opsteegh, M.; Langelaar, M.; Sprong, H.; Den Hartog, L.; De Craeye, S.; Bokken, G.; Ajzenberg, D.; Kijlstra, A.; Van der Giessen, J. Direct detection and genotyping of Toxoplasma gondii in meat samples using magnetic capture and PCR. Int. J. Food Microbiol. 2010, 139, 193–201. [Google Scholar] [CrossRef]
- Wolffs, P.; Norling, B.; Rådström, P. Risk assessment of false-positive quantitative real-time PCR results in food, due to detection of DNA originating from dead cells. J. Microbiol. Methods 2005, 60, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Salipante, S.J.; Jerome, K.R. Digital PCR—An emerging technology with broad applications in microbiology. Clin. Chem. 2020, 66, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.; Jeong, S.W.; Bae, N.H.; Song, Y.; Lee, T.J.; Lee, M.K.; Lee, S.J.; Lee, K.G. Droplet-based digital PCR system for detection of single-cell level of foodborne pathogens. BioChip J. 2017, 11, 329–337. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Rapid Increase of a SARS-CoV-2 Variant with Multiple Spike Protein Mutations Observed in the United Kingdom; ECDC: Stockholm, Sweden, 20 December 2020. [Google Scholar]
- Whittaker, P.; Fry, F.S.; Curtis, S.K.; Al-Khaldi, S.F.; Mossoba, M.M.; Yurawecz, M.P.; Dunkel, V.C. Use of fatty acid profiles to identify food-borne bacterial pathogens and aerobic endospore-forming bacilli. J. Agric. Food Chem. 2005, 53, 3735–3742. [Google Scholar] [CrossRef]
- Pinu, F.R. Early detection of food pathogens and food spoilage microorganisms: Application of metabolomics. Trends Food Sci. Technol. 2016, 54, 213–215. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, S.; Pu, Q.; Li, Y.; Wang, X.; Jiang, Y.; Yang, D.; Yang, Y.; Yang, J.; Sun, C. Rapid identification of Staphylococcus aureus, Vibrio parahaemolyticus and Shigella sonnei in foods by solid phase microextraction coupled with gas chromatography–mass spectrometry. Food Chem. 2018, 262, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Harris, G.A.; Galhena, A.S.; Fernández, F.M. Ambient Sampling/Ionization Mass Spectrometry: Applications and Current Trends. Anal. Chem. 2011, 83, 4508–4538. [Google Scholar] [CrossRef]
- Rodríguez-Lorenzo, L.; Garrido-Maestu, A.; Bhunia, A.K.; Espiña, B.; Prado, M.; Dieǵuez, L.; Abalde-Cela, S. Gold nanostars for the detection of foodborne pathogens via surface-enhanced Raman scattering combined with microfluidics. ACS Appl. Nano Mater. 2019, 2, 6081–6086. [Google Scholar] [CrossRef]
- American Association of Veterinary Laboratory Diagnosticians. Available online: http://www.aavld.org (accessed on 28 January 2021).
- Collaborating Veterinary Laboratories. Available online: http://www.covetlab.org (accessed on 28 January 2021).
- European Association of Veterinary Laboratory Diagnosticians. Available online: http://www.eavld.org (accessed on 28 January 2021).
- World Organization for Animal Health. Principles of validation of diagnostic assays for infectious diseases. In OIE Manual of Diagnostic Tests and Vaccines for Terrestrial Animals (Mammals, Birds and Bees), 6th ed.; OIE Biological Standards Commission, Ed.; Office International Des Epizooties: Paris, France, 2008; Volume 1, Chapter 1.1.4; pp. 34–35. [Google Scholar]
- Wild, D.; Sheehan, C. Standardization and calibration. In The Immunoassay Handbook, 4th ed.; Wild, D., Ed.; Elsevier: Amsterdam, The Netherlands, 2013; pp. 315–322. [Google Scholar] [CrossRef]
- Cardoso, J.R.; Pereira, L.M.; Iversen, M.D.; Ramos, A.L. What is gold standard and what is ground truth? Dent. Press J. Orthod. 2014, 19, 27–30. [Google Scholar] [CrossRef] [Green Version]
- Cundell, T. The limitations of the colony-forming unit in microbiology. Eur. Pharm. Rev. 2015, 20, 11–13. [Google Scholar]
- Tang, S.; Orsi, R.H.; Luo, H.; Ge, C.; Zhang, G.; Baker, R.C.; Stevenson, A.; Wiedmann, M. Assessment and comparison of molecular subtyping and characterization methods for Salmonella. Front. Microbiol. 2019, 10, 1591. [Google Scholar] [CrossRef] [Green Version]
- Azadeh, M.; Sondag, P.; Wang, Y.; Raines, M.; Sailstad, J. Quality controls in ligand binding assays: Recommendations and best practices for preparation, qualification, maintenance of lot to lot consistency, and prevention of assay drift. AAPS J. 2019, 21, 89. [Google Scholar] [CrossRef] [Green Version]
- Bonardi, S. Salmonella in the pork production chain and its impact on human health in the European Union. Epidemiol. Infect. 2017, 145, 1513–1526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belœil, P.A.; Chauvin, C.; Proux, K.; Rose, N.; Queguiner, S.; Eveno, E.; Houdayer, C.; Rose, V.; Fravalo, P.; Madec, F. Longitudinal serological responses to Salmonella enterica of growing pigs in a subclinically infected herd. Prev. Vet. Med. 2003, 60, 207–226. [Google Scholar] [CrossRef]
- Berends, B.R.; Urlings, H.A.P.; Snijders, J.M.A.; Van Knapen, F. Identification and quantification of risk factors in animal management and transport regarding Salmonella spp. in pigs. Int. J. Food Microbiol. 1996, 30, 37–53. [Google Scholar] [CrossRef]
- Lo, F.; Wong, D.M.A.; Hald, T. (Eds.) Salmonella in Pork (SALINPORK): Pre-Harvest and Harvest Control Options Based on Epidemiologic, Diagnostic and Economic Research; Contract No: FAIR1 CT95-0400. Copenhagen, Denmark, 2002. Available online: https://cordis.europa.eu/project/id/FAIR950400 (accessed on 28 January 2021).
- Baggesen, D.L.; Wegener, H.C.; Bager, F.; Stege, H.; Christensen, J. Herd prevalence of Salmonella enterica infections in Danish slaughter pigs determined by microbiological testing. Prev. Vet. Med. 1996, 26, 201–213. [Google Scholar] [CrossRef]
- Nollet, N.; Maes, D.; Duchateau, L.; Hautekiet, V.; Houf, K.; Van Hoof, J.; De Zutter, L.; De Kruif, A.; Geers, R. Discrepancies between the isolation of Salmonella from mesenteric lymph nodes and the results of serological screening in slaughter pigs. Vet. Res. 2005, 36, 545–555. [Google Scholar] [CrossRef] [Green Version]
- Farzan, A.; Friendship, R.; Dewey, C. Evaluation of enzyme-linked immunosorbent assay (ELISA) tests and culture for determining Salmonella status of a pig herd. Epidemiol. Infect. 2007, 135, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, E.; Hilbert, F.; Paulsen, P.; Smulders, F.J.M.; Rossmanith, W. Salmonella diagnosis in pig production: Methodological problems in monitoring the prevalence in pigs and pork. J. Food Prot. 2007, 70, 1246–1248. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority Task Force on Zoonoses. Report of the Task Force on Zoonoses Data Collection on the analysis of the baseline survey on the prevalence of Salmonella in slaughter pigs, in the EU, 2006–2007. Part A: Salmonella prevalence estimates. EFSA J. 2008, 135, 1–111. [Google Scholar]
- Swanenburg, M.; Berends, B.R.; Urlings, H.A.; Snijders, J.M.; Van Knapen, F. Epidemiological investigations into the sources of Salmonella contamination of pork. Berl. Munch. Tierarztl. Wochenschr. 2001, 114, 356–359. [Google Scholar]
- Mainar-Jaime, R.C.; Casanova-Higes, A.; Andrés-Barranco, S.; Vico, J.P. Looking for new approaches for the use of serology in the context of control programmes against pig salmonellosis. Zoonoses Public Health 2018, 65, e222–e228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felin, E.; Hälli, O.; Heinonen, M.; Jukola, E.; Fredriksson-Ahomaa, M. Assessment of the feasibility of serological monitoring and on-farm information about health status for the future meat inspection of fattening pigs. Prev. Vet. Med. 2019, 162, 76–82. [Google Scholar] [CrossRef] [Green Version]
- Snary, E.L.; Munday, D.K.; Arnold, M.E.; Cook, A.J.C. Zoonoses action plan Salmonella monitoring programme: An investigation of the sampling protocol. J. Food Prot. 2010, 73, 488–494. [Google Scholar] [CrossRef]
- Hanssen, E.J.M.; Swanenburg, M.; Maassen, C.B.M. The Dutch Salmonella Monitoring Programme for Pigs and Some Recommendations for Control Plans in the Future. In Proceedings of the 7th International Symposium on the Epidemiology & Control of Foodborne Pathogens in Pork, Verona, Italy, 9–11 May 2007; pp. 169–172. [Google Scholar] [CrossRef]
- Wegener, H.C.; Hald, T.; Wong, L.F.; Madsen, M.; Korsgaard, H.; Bager, F.; Gerner-Smidt, P.; Mølbak, K. Salmonella control programs in Denmark. Emerg. Infect. Dis. 2003, 9, 774–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berends, B.R.; Snijders, J.M.; Van Logtestijn, J.G. Efficacy of current EC meat inspection procedures and some proposed revisions with respect to microbiological safety: A critical review. Vet. Rec. 1993, 133, 411–415. [Google Scholar] [CrossRef]
- Snijders, J.M.A.; Van Knapen, F. Prevention of human diseases by an integrated quality control system. Livest. Prod. Sci. 2002, 76, 203–206. [Google Scholar] [CrossRef]
- Robert, M.K. Memorium. Crit. Rev. Toxicol. 2007, 37, 627. [Google Scholar] [CrossRef]
- Kahn, L.H. Perspective: The one-health way. Nature 2017, 543, S47. [Google Scholar] [CrossRef]
- Van Knapen, F. Veterinary public health: Past, present, and future. Vet. Q. 2000, 22, 61–62. [Google Scholar] [CrossRef] [PubMed]
- Riess, L.E.; Hoelzer, K. Implementation of visual-only swine inspection in the European Union: Challenges, opportunities, and lessons learned. J. Food Prot. 2020, 83, 1918–1928. [Google Scholar] [CrossRef]
- Messens, W.; Hempen, M.; Koutsoumanis, K. Use of predictive modelling in recent work of the Panel on Biological Hazards of the European Food Safety Authority. Microb. Risk Anal. 2018, 10, 37–43. [Google Scholar] [CrossRef]
- McMeekin, T.; JBowman, J.; McQuestin, O.; Mellefont, L.; Ross, T.; Tamplin, M. The future of predictive microbiology: Strategic research, innovative applications and great expectations. Int. J. Food Microbiol. 2008, 128, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Bergwerff, A.A. Rapid assays for detection of residus of veterinary drugs. In Rapid Methods for Biological and Chemical Contaminants in Food and Feed; Van Amerongen, A., Barug, D., Lauwaars, M., Eds.; Wageningen Academic Publishers: Wageningen, The Netherlands, 2005; pp. 259–292. [Google Scholar]
- Commission of The European Communities. Commission Regulation (EC) No 2075/2005 of 5 December 2005 laying down specific rules on official controls for Trichinella in meat (consolidated version including Commission Regulation amendments and corrections). Off. J. Eur. Union 2005, L338, 60–82. [Google Scholar]
- Pozio, E. World distribution of Trichinella spp. infections in animals and humans. Vet. Parasitol. 2007, 149, 3–21. [Google Scholar] [CrossRef]
- Bokken, G.C.A.M.; Van Eerden, E.; Opsteegh, M.; Augustijn, M.; Graat, E.A.M.; Franssen, F.F.J.; Görlich, K.; Buschtöns, S.; Tenter, A.M.; Van der Giessen, J.W.B.; et al. Specific serum antibody responses following a Toxoplasma gondii and Trichinella spiralis co-infection in swine. Vet. Parasitol. 2012, 184, 126–132. [Google Scholar] [CrossRef]
- Yousefi, H.; Su, H.-M.; Imani, S.M.; Alkhaldi, K.; Filipe, C.D.M.; Didar, T.F. Intelligent food packaging: A review of smart sensing technologies for monitoring food quality. ACS Sens. 2019, 4, 808–821. [Google Scholar] [CrossRef]
- Friesema, I.H.M.; De Boer, R.F.; Duizer, E.; Kortbeek, L.M.; Notermans, D.W.; Smeulders, A.; Bogerman, J.; Pronk, M.J.H.; Uil, J.J.; BrInkman, K.; et al. Aetiology of acute gastroenteritis in adults requiring hospitalization in The Netherlands. Epidemiol. Infect. 2012, 140, 1780–1786. [Google Scholar] [CrossRef]
- Jansen, A.; Stark, K.; Kunkel, J.; Schreier, E.; Ignatius, R.; Liesenfeld, O.; Werber, D.; Göbel, U.B.; Zeitz, M.; Schneider, T. Aetiology of community-acquired, acute gastroenteritis in hospitalised adults: A prospective cohort study. BMC Infect. Dis. 2008, 8, 143. [Google Scholar] [CrossRef] [Green Version]
- Thissen, J.B.; Be, N.A.; McLoughlin, K.; Gardner, S.; Rack, P.G.; Shapero, M.H.; Rowland, R.R.R.; Slezak, T.; Jaing, C.J. Axiom Microbiome Array, the next generation microarray for high-throughput pathogen and microbiome analysis. PLoS ONE 2019, 14, e0212045. [Google Scholar] [CrossRef] [Green Version]
- Strategic Consulting. Microbiology Testing in the Global Food Industry, 8th ed. 2013. Available online: https://www.strategic-consult.com/product/food-micro-eighth-edition-microbiology-testing-global-food-industry/ (accessed on 23 January 2021).
- Bolton, D.J. Campylobacter virulence and survival factors. Food Microbiol. 2015, 48, 99–108. [Google Scholar] [CrossRef] [PubMed]
- An, X.; Zuo, P.; Ye, B.-C. A single cell droplet microfluidic system for quantitative determination of food-borne pathogens. Talanta 2020, 209, 120571. [Google Scholar] [CrossRef] [PubMed]
- Moschou, D. How close are we to a real Star Trek-style medical tricorder? Conversation. 16 June 2017. Available online: https://theconversation.com/how-close-are-we-to-a-real-star-trek-style-medical-tricorder-77977 (accessed on 11 December 2020).






| Pathogenic Micro-Organism | Confirmed Human Cases (Number) | Case Fatality (%) |
|---|---|---|
| Campylobacter | 220,682 | 0.03 |
| Salmonella | 87,923 | 0.22 |
| Shiga toxin-producing E. coli (STEC) | 7775 | 0.21 |
| Yersinia | 6961 | 0.05 |
| Listeria | 2621 | 17.6 |
| Step/Phase | Note |
|---|---|
| To investigate | Water: environment, processing water, drinking water (for animals or to prepare a meal). Environment: farm (including wild animals and insects in its surroundings), processing plant, abattoir, butcher, greengrocer, kitchen, etc. Pre-products: carcass, ingredients (herbs), etc. Products: fruit, meat, sliced vegetables, ready-to-eat, salads, etc. |
| Sampling | Sample quality and size should reflect what is investigated (food, flock, farm, herd, retailer, kitchen, etc.) |
| Transport | Identifiable at all times, properly cooled, and with no risk of cross-contamination |
| Sample treatment | Acclimatization Homogenize if required (as a matter of fact, homogenization is a specialism in itself) |
| Pre-enrichment | Resuscitate and proliferate bacteria to determine even low numbers |
| Selective enrichment | Proliferate the aimed pathogen exclusively |
| Culture evaluation | Gauge selective culture by assessing color, smell, turbidity and microscopical investigation, Gram staining, etc. |
| Analysis | Traditional plating, agglutination, enzymic assays, LFD, (IMS) ELISA, apta-/geno-/immune-/phagosensors, LoaC, LoaD, nucleic acid sequence assays |
| Verification | Confirmation and identification |
| Pathogen/Predictor | Food Product/Matrix | Norm |
|---|---|---|
| Listeria monocytogenes | Ready-to-eat a | Absent in 25 g |
| Salmonella | Cheese, butter, cream from raw or non-pasteurized milk | Absent in 25 g |
| Salmonella | Meat products intended to be eaten raw | Absent in 25 g |
| Salmonella | Meat preparations intended to be eaten cooked b | Absent in 10 g |
| Staphylococcal enterotoxins | Cheeses, milk powder, and whey powder | not detected in 25 g c |
| Enterobacteriaceae | Egg products | 10 or 100 CFU/mL or CFU/g |
| Campylobacter spp. | Carcasses of broilers | 1000 CFU/g |
| STEC O157, O26, O111, O103, O145 and O104:H4 | sprouts | not detected in 25 g c |
| Requirement | Note |
|---|---|
| Low cost | Test outcome should provide sufficient information to make a contemplated decision at costs that balances investments and consequences. Costs include overhead, maintenance, personnel, laboratory footprint, disposables, etc. |
| One-pot reaction | Complete preparation and processing in a single vial/tube. No addition of reagents required (~ easy-to-use). |
| Range of application/ operation | Versatile: sensu lato applicable to fresh and processed food matrices, and/or for samples from feed, plants, and animals. Suitable for all relevant bioagents. |
| Multi-analyte/multiplex | Able to determine multiple targets in a single analysis run. |
| High-throughput | Multiple samples run simultaneously. |
| (relative) accuracy | Free from systematic and random errors. |
| Reproducible (precise) | No or insignificant variation (in-)between runs, days, machines, analysts, etc. |
| Repeatable | No or insignificant variation between laboratories, batches, lots. |
| (Relative) specificity | 100% specific compared to the reference method (no false positives); able to distinguish at the strain level. |
| (Relative) sensitivity | 100% sensitive compared to the reference method (no false negatives); able to detect (viable) MO at required sensitivity a. Excellent signal-to-noise ratio. |
| Confirmation of analyte | Juridical problems arise when the identity of the analyte is not confirmed. There should be a reliable answer to the question: what is the degree of certainty of the identity of the aimed target giving the result? |
| Automable | A stand-alone instrument with limited hands-on time. The frequency of personnel attention is low. |
| Portable/Point-of-care (PoC) | Ambulant, performing analysis in/at/on-line of the food-chain with interpretable results available on site. |
| User-friendly and fail-safe | Easy-to-use, performed by non-skilled personnel, preventing facultative and inherent errors. In other words, the assay is rugged and gives the same result even when test conditions such as incubation times, operator, pH, reagent concentrations, temperature, etc. vary. This also includes safe to use. |
| Easy to interpret | Analysis data give a transparent, unambiguous, and understandable result |
| Instantaneous result | Results available real- or near-time. Although not meaning the same, time needed for the whole analytical process from sample collection to result (TTR) should be short. In practice, TTR can imply a result within a working day/before the next phase in the food-chain. |
| No sample treatment | The test portion is obtained directly from the laboratory sample. |
| Robust | Reliable operation under e.g., humid, varying temperatures, bumping conditions. No drift/long-term stability. |
| High | Antes a | Drivers b |
|---|---|---|
| Relevance |
|
|
| Low | Neutrals c | Fool’s Gold d |
|
| |
| Low Differentiation High | ||
| Traditional (Direct Antigen) | Alternative (Indirect Antigen) |
|---|---|
| Laboratory-bound | Point-of-care possible |
| Intensive material use | Relative expensive material |
| TTR long | TTR usually short |
| Reproducibility moderate | Reproducibility mostly better |
| Almost no instruments | Needs more devices |
| Difficult to automate | Possibilities to automate |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bergwerff, A.A.; Debast, S.B. Modernization of Control of Pathogenic Micro-Organisms in the Food-Chain Requires a Durable Role for Immunoaffinity-Based Detection Methodology—A Review. Foods 2021, 10, 832. https://doi.org/10.3390/foods10040832
Bergwerff AA, Debast SB. Modernization of Control of Pathogenic Micro-Organisms in the Food-Chain Requires a Durable Role for Immunoaffinity-Based Detection Methodology—A Review. Foods. 2021; 10(4):832. https://doi.org/10.3390/foods10040832
Chicago/Turabian StyleBergwerff, Aldert A., and Sylvia B. Debast. 2021. "Modernization of Control of Pathogenic Micro-Organisms in the Food-Chain Requires a Durable Role for Immunoaffinity-Based Detection Methodology—A Review" Foods 10, no. 4: 832. https://doi.org/10.3390/foods10040832
APA StyleBergwerff, A. A., & Debast, S. B. (2021). Modernization of Control of Pathogenic Micro-Organisms in the Food-Chain Requires a Durable Role for Immunoaffinity-Based Detection Methodology—A Review. Foods, 10(4), 832. https://doi.org/10.3390/foods10040832