Extending the Lipidome Coverage by Combining Different Mass Spectrometric Platforms: An Innovative Strategy to Answer Chemical Food Safety Issues
Abstract
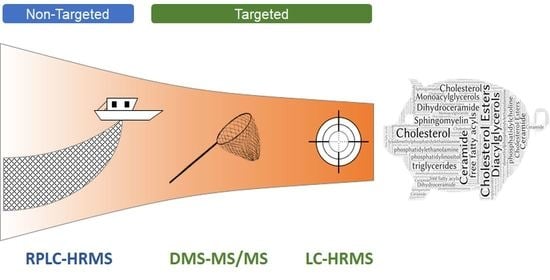
:1. Introduction
2. Materials and Methods
2.1. Animal Experiment
2.2. Analytical Platforms
2.3. Data Analysis
3. Results
3.1. Non-Targeted RPLC-HRMS
3.2. Lipidyzer™ Platform
3.3. TG Platform
4. Discussion
4.1. Assessment of the Complementarity between Platforms
- From the non-targeted method, TGs were annotated from their three FA chains (e.g., “TG(16:0_17:0_18:1)”), based on the annotation results from LipidSearch after data-dependent MS/MS. Although allowing confident assignment, the results of such an approach may in some particular cases be considered with caution as illustrated hereafter. Among the selected features, for instance, some lipids (M926T1080 and M921T1080; highlighted in light grey as well as M898T1065 and M893T1066 highlighted in dark grey in Table 2) were annotated as adducts of the same TG. These features were initially not discarded during the data processing step because of an inconsistency between the adduct annotation between the CAMERA package and LipidSearch. In addition, two other features (M919T1066 and M924T1066; highlighted in blue in Table 2) were annotated as two different TG when they could potentially be two adducts of the same lipid as they are isomers of TG(55:2).
- In Lipidyzer™, TG results were expressed with the shorthand annotation nomenclature (total number of carbons and unsaturations among the three FA chains and the precision on one of them), such as ”TG51:1-FA16:0”. While technically correct, this leads to an overestimation of the TG, as previously highlighted in the literature [31]. Moreover, several Lipidyzer™ candidates (e.g., TG51:1-FA18:1 and TG51:1-FA16:0) can correspond to a single TG feature in RP LC-HRMS (e.g., TG(16:0_17:0_18:1)), and vice-versa, thus complicating result comparison.
- Because of previous issues in TG assignment, a dedicated platform for the determination of TG regioisomeric composition was used [35]. It is interesting to note that the TGs highlighted with the dedicated tool were not those annotated in non-targeted data. Moreover, after conversion to the corresponding shorthand annotation to allow such a comparison, none of them was deemed as significant with Lipidyzer™, which could be due to the overestimation of TG with the latter. Conversely, none of the discriminant TG highlighted within the RPLC-HRMS results were monitored with the TG platform since it is designed for the analysis of even FA chains TG only. It is interesting to note that this specific platform allowed obtaining confident results on TG and the position (sn-1(3) versus sn-2) of their constituting FA chains. Thus, it yielded finer results than the combined use of non-targeted and Lipidyzer platforms—an approach that was already explored by Contrepois et al. [31].
4.2. Biological Interpretation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Council Directive 88/146/EEC. Council Directive 88/146/EEC Prohibiting the Use Livestock Farming of Certain Substances Having a Hormonal Action. Available online: http://data.europa.eu/eli/dir/1988/146/oj (accessed on 25 May 2021).
- European Parliament and Council. Regulation (EU) 2017/625 on Official Controls and Other Official Activities Performed to Ensure the Application of Food and Feed Law, Rules on Animal Health and Welfare, Plant Health and Plant Protection Products, Amending Regulations (EC) No 999/2001, (EC) No 396/2005, (EC) No 1069/2009, (EC) No 1107/2009, (EU) No 1151/2012, (EU) No 652/2014, (EU) 2016/429 and (EU) 2016/2031 of the European Parliament and of the Council, Council Regulations (EC) No 1/2005 and (EC) No 1099/2009 and Council Directives 98/58/EC, 1999/74/EC, 2007/43/EC, 2008/119/EC and 2008/120/EC, and repealing Regulations (EC) No 854/2004 and (EC) No 882/2004 of the European Parliament and of the Council, Council Directives 89/608/EEC, 89/662/EEC, 90/425/EEC, 91/496/EEC, 96/23/EC, 96/93/EC and 97/78/EC and Council Decision 92/438/EEC (Official Controls Regulation). OJ L 95, 7.4. 2017, pp. 1–142. Available online: http://data.europa.eu/eli/reg/2017/625/oj (accessed on 25 May 2021).
- Pinel, G.; Weigel, S.; Antignac, J.-P.; Mooney, M.; Elliott, C.; Nielen, M.; Le Bizec, B. Targeted and untargeted profiling of biological fluids to screen for anabolic practices in cattle. TrAC Trends Anal. Chem. 2010, 29, 1269–1280. [Google Scholar] [CrossRef]
- Gallart-Ayala, H.; Chéreau, S.; Dervilly-Pinel, G.; Le Bizec, B. Potential of mass spectrometry metabolomics for chemical food safety. Bioanalysis 2015, 7, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Courant, F.; Pinel, G.; Bichon, E.; Monteau, F.; Antignac, J.-P.; Le Bizec, B. Development of a metabolomic approach based on liquid chromatography-high resolution mass spectrometry to screen for clenbuterol abuse in calves. Analyst 2009, 134, 1637–1646. [Google Scholar] [CrossRef] [PubMed]
- Stella, R.; Dervilly-Pinel, G.; Bovo, D.; Mastrorilli, E.; Royer, A.-L.; Angeletti, R.; Le Bizec, B.; Biancotto, G. Metabolomics analysis of liver reveals profile disruption in bovines upon steroid treatment. Metabolomics 2017, 13, 80. [Google Scholar] [CrossRef]
- Lu, H.; Zhang, H.; Zhu, T.; Xiao, Y.; Xie, S.; Gu, H.; Cui, M.; Luo, L. Metabolic Effects of Clenbuterol and Salbutamol on Pork Meat Studied Using Internal Extractive Electrospray Ionization Mass Spectrometry. Sci. Rep. 2017, 7, 5136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dervilly-Pinel, G.; Courant, F.; Chéreau, S.; Royer, A.-L.; Boyard-Kieken, F.; Antignac, J.-P.; Monteau, F.; Le Bizec, B. Metabolomics in food analysis: Application to the control of forbidden substances. Drug Test. Anal. 2012, 4, 59–69. [Google Scholar] [CrossRef]
- Dervilly-Pinel, G.; Weigel, S.; Lommen, A.; Chereau, S.; Rambaud, L.; Essers, M.; Antignac, J.-P.; Nielen, M.W.; Le Bizec, B. Assessment of two complementary liquid chromatography coupled to high resolution mass spectrometry metabolomics strategies for the screening of anabolic steroid treatment in calves. Anal. Chim. Acta 2011, 700, 144–154. [Google Scholar] [CrossRef]
- Jacob, C.C.; Dervilly-Pinel, G.; Biancotto, G.; Monteau, F.; Le Bizec, B. Global urine fingerprinting by LC-ESI(+)-HRMS for better characterization of metabolic pathway disruption upon anabolic practices in bovine. Metabolomics 2014, 11, 184–197. [Google Scholar] [CrossRef]
- Nzoughet, J.J.K.; Dervilly-Pinel, G.; Chéreau, S.; Biancotto, G.; Monteau, F.; Elliott, C.T.; Le Bizec, B. First insights into serum metabolomics of trenbolone/estradiol implanted bovines; screening model to predict hormone-treated and control animals’ status. Metabolomics 2015, 11, 1184–1196. [Google Scholar] [CrossRef]
- Nzoughet, J.K.; Gallart-Ayala, H.; Biancotto, G.; Hennig, K.; Dervilly-Pinel, G.; Le Bizec, B. Hydrophilic interaction (HILIC) and reverse phase liquid chromatography (RPLC)–high resolution MS for characterizing lipids profile disruption in serum of anabolic implanted bovines. Metabolomics 2015, 11, 1884–1895. [Google Scholar] [CrossRef]
- Guitton, Y.; Dervilly-Pinel, G.; Jandova, R.; Stead, S.; Takats, Z.; Le Bizec, B. Rapid evaporative ionisation mass spectrometry and chemometrics for high-throughput screening of growth promoters in meat producing animals. Food Addit. Contam. Part A 2018, 35, 900–910. [Google Scholar] [CrossRef] [PubMed]
- Wenk, M.R. Lipidomics: New Tools and Applications. Cell 2010, 143, 888–895. [Google Scholar] [CrossRef] [Green Version]
- Ryan, E.; Reid, G.E. Chemical Derivatization and Ultrahigh Resolution and Accurate Mass Spectrometry Strategies for “Shotgun” Lipidome Analysis. Accounts Chem. Res. 2016, 49, 1596–1604. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.-Y.; Wu, S.-P.; Liu, S.; Zhang, Y.; Lin, R.-C. Ultra-performance liquid chromatography–mass spectrometry as a sensitive and powerful technology in lipidomic applications. Chem. Interact. 2014, 220, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yang, L.; Bai, Y.; Liu, H. Analytical Methods in Lipidomics and Their Applications. Anal. Chem. 2014, 86, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [Green Version]
- Folch, J.; Ascoli, I.; Lees, M.; Meath, J.; LeBaron, F. Preparation of lipide extracts from brain tissue. J. Biol. Chem. 1951, 191, 833–841. [Google Scholar] [CrossRef]
- Tumanov, S.; Kamphorst, J.J. Recent advances in expanding the coverage of the lipidome. Curr. Opin. Biotechnol. 2017, 43, 127–133. [Google Scholar] [CrossRef]
- Yang, K.; Han, X. Lipidomics: Techniques, Applications, and Outcomes Related to Biomedical Sciences. Trends Biochem. Sci. 2016, 41, 954–969. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.; van der Heijden, R.; Wang, M.; van der Greef, J.; Hankemeier, T.; Xu, G. Analytical strategies in lipidomics and applications in disease biomarker discovery. J. Chromatogr. B 2009, 877, 2836–2846. [Google Scholar] [CrossRef]
- Triebl, A.; Hartler, J.; Trötzmüller, M.; Köfeler, H.C. Lipidomics: Prospects from a technological perspective. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2017, 1862, 740–746. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Gross, R.W. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: A bridge to lipidomics. J. Lipid Res. 2003, 44, 1071–1079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwudke, D.; Oegema, J.; Burton, L.; Entchev, E.; Hannich, J.T.; Ejsing, C.S.; Kurzchalia, T.; Shevchenko, A. Lipid Profiling by Multiple Precursor and Neutral Loss Scanning Driven by the Data-Dependent Acquisition. Anal. Chem. 2006, 78, 585–595. [Google Scholar] [CrossRef]
- Schwudke, D.; Schuhmann, K.; Herzog, R.; Bornstein, S.R.; Shevchenko, A. Shotgun Lipidomics on High Resolution Mass Spectrometers. Cold Spring Harb. Perspect. Biol. 2011, 3, a004614. [Google Scholar] [CrossRef] [Green Version]
- Almeida, R.; Pauling, J.K.; Sokol, E.; Hannibal-Bach, H.K.; Ejsing, C.S. Comprehensive Lipidome Analysis by Shotgun Lipidomics on a Hybrid Quadrupole-Orbitrap-Linear Ion Trap Mass Spectrometer. J. Am. Soc. Mass Spectrom. 2014, 26, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Hinz, C.; Liggi, S.; Griffin, J.L. The potential of Ion Mobility Mass Spectrometry for high-throughput and high-resolution lipidomics. Curr. Opin. Chem. Biol. 2018, 42, 42–50. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.-C.; Yokomizo, T. Applications of mass spectrometry-based targeted and non-targeted lipidomics. Biochem. Biophys. Res. Commun. 2018, 504, 576–581. [Google Scholar] [CrossRef]
- Cajka, T.; Fiehn, O. Toward Merging Untargeted and Targeted Methods in Mass Spectrometry-Based Metabolomics and Lipidomics. Anal. Chem. 2016, 88, 524–545. [Google Scholar] [CrossRef]
- Contrepois, K.; Mahmoudi, S.; Ubhi, B.K.; Papsdorf, K.; Hornburg, D.; Brunet, A.; Snyder, M. Cross-Platform Comparison of Untargeted and Targeted Lipidomics Approaches on Aging Mouse Plasma. Sci. Rep. 2018, 8, 17747. [Google Scholar] [CrossRef]
- Peng, T.; Royer, A.-L.; Guitton, Y.; Le Bizec, B.; Dervilly-Pinel, G. Serum-based metabolomics characterization of pigs treated with ractopamine. Metabolomics 2017, 13, 77. [Google Scholar] [CrossRef]
- Lintonen, T.P.I.; Baker, P.R.S.; Suoniemi, M.; Ubhi, B.K.; Koistinen, K.M.; Duchoslav, E.; Campbell, J.L.; Ekroos, K. Differential Mobility Spectrometry-Driven Shotgun Lipidomics. Anal. Chem. 2014, 86, 9662–9669. [Google Scholar] [CrossRef]
- Ubhi, B.K.; Conner, A.; Duchoslav, E.; Evans, A.; Robinson, R.; Wang, L.; Baker, P.R.; Watkins, S. A Novel Lipid Screening Platform that Provides a Complete Solution for Lipidomics Research; Sciex: Vaughan, ON, Canada, 2016; pp. 1–4. [Google Scholar]
- Balgoma, D.; Guitton, Y.; Evans, J.J.; Le Bizec, B.; Dervilly-Pinel, G.; Meynier, A.; David, B.; Yann, G.; Jason, J.E.; Bruno, L.B.; et al. Modeling the fragmentation patterns of triacylglycerides in mass spectrometry allows the quantification of the regioisomers with a minimal number of standards. Anal. Chim. Acta 2019, 1057, 60–69. [Google Scholar] [CrossRef]
- Kessner, D.; Chambers, M.; Burke, R.; Agus, D.; Mallick, P. ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics 2008, 24, 2534–2536. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Van Der Kloet, F.M.; Bobeldijk, I.; Verheij, E.R.; Jellema, R. Analytical Error Reduction Using Single Point Calibration for Accurate and Precise Metabolomic Phenotyping. J. Proteome Res. 2009, 8, 5132–5141. [Google Scholar] [CrossRef] [PubMed]
- Marchand, J.; Martineau, E.; Guitton, Y.; Le Bizec, B.; Dervilly-Pinel, G.; Giraudeau, P. A multidimensional 1H NMR lipidomics workflow to address chemical food safety issues. Metabolomics 2018, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Navas-Iglesias, N.; Carrasco-Pancorbo, A.; Cuadros-Rodríguez, L. From lipids analysis towards lipidomics, a new challenge for the analytical chemistry of the 21st century. Part II: Analytical lipidomics. TrAC Trends Anal. Chem. 2009, 28, 393–403. [Google Scholar] [CrossRef]
- Eriksson, L.; Trygg, J.; Wold, S. CV-ANOVA for significance testing of PLS and OPLS® models. J. Chemom. 2008, 22, 594–600. [Google Scholar] [CrossRef]
- Dervilly-Pinel, G.; Chereau, S.; Cesbron, N.; Monteau, F.; Le Bizec, B. LC-HRMS based metabolomics screening model to detect various β-agonists treatments in bovines. Metabolomics 2015, 11, 403–411. [Google Scholar] [CrossRef]
- Galindo-Prieto, B.; Eriksson, L.; Trygg, J. Variable influence on projection (VIP) for orthogonal projections to latent structures (OPLS). J. Chemom. 2014, 28, 623–632. [Google Scholar] [CrossRef]
- Giacomoni, F.; Le Corguillé, G.; Monsoor, M.; Landi, M.; Pericard, P.; Pétéra, M.; Duperier, C.; Tremblay-Franco, M.; Martin, J.-F.; Jacob, D.; et al. Workflow4Metabolomics: A collaborative research infrastructure for computational metabolomics. Bioinformatics 2015, 31, 1493–1495. [Google Scholar] [CrossRef] [Green Version]
- Guitton, Y.; Tremblay-Franco, M.; Le Corguillé, G.; Martin, J.-F.; Pétéra, M.; Roger-Mele, P.; Delabrière, A.; Goulitquer, S.; Monsoor, M.; Duperier, C.; et al. Create, run, share, publish, and reference your LC–MS, FIA–MS, GC–MS, and NMR data analysis workflows with the Workflow4Metabolomics 3.0 Galaxy online infrastructure for metabolomics. Int. J. Biochem. Cell Biol. 2017, 93, 89–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Campbell, J.L.; Le Blanc, J.C.Y.; Baker, P.R. Structural identification of triacylglycerol isomers using electron impact excitation of ions from organics (EIEIO). J. Lipid Res. 2016, 57, 2015–2027. [Google Scholar] [CrossRef] [Green Version]
- Nagy, K.; Sandoz, L.; Destaillats, F.; Schafer, O. Mapping the regioisomeric distribution of fatty acids in triacylglycerols by hybrid mass spectrometry. J. Lipid Res. 2013, 54, 290–305. [Google Scholar] [CrossRef] [Green Version]
- Bird, S.S.; Marur, V.R.; Sniatynski, M.J.; Greenberg, H.K.; Kristal, B.S. Serum Lipidomics Profiling Using LC–MS and High-Energy Collisional Dissociation Fragmentation: Focus on Triglyceride Detection and Characterization. Anal. Chem. 2011, 83, 6648–6657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- E Watkins, L.; Jones, D.J.; Mowrey, D.H.; Anderson, D.B.; Veenhuizen, E.L. The effect of various levels of ractopamine hydrochloride on the performance and carcass characteristics of finishing swine. J. Anim. Sci. 1990, 68, 3588–3595. [Google Scholar] [CrossRef]
- Council Directive 96/22/EC. Council Directive 96/22/EC of 29 April 1996 Concerning the Prohibition on the Use in Stockfarming of Certain Substances Having a Hormonal or Thyrostatic Action and of Beta-Agonists, and Repealing Directives 81/602/EEC, 88/146/EEC and 88/299/EEC. 1996. Available online: http://data.europa.eu/eli/dir/1996/22/oj (accessed on 25 May 2021).
- Dunshea, F.R. Effect of metabolism modifiers on lipid metabolism in the pig. J. Anim. Sci. 1993, 71, 1966–1977. [Google Scholar] [CrossRef] [PubMed]
- Dunshea, F.R.; Leur, B.J.; Tilbrook, A.J.; King, R.H. Ractopamine increases glucose turnover without affecting lipogenesis in the pig. Aust. J. Agric. Res. 1998, 49, 1147. [Google Scholar] [CrossRef]
- Ferreira, M.S.D.S.; Garbossa, C.A.P.; Oberlender, G.; Pereira, L.J.; Zangeronimo, M.G.; De Sousa, R.V.; Cantarelli, V.D.S. Effect of ractopamine on lipid metabolism in vivo—A systematic review. Braz. Arch. Biol. Technol. 2013, 56, 35–43. [Google Scholar] [CrossRef]
- Paris, A.; André, F.; Antignac, J.P.; Bonneau, M.; Briant, C.; Caraty, A.; Chilliard, Y.; Cognié, Y.; Combarnous, Y.; Cravedi, J.P.; et al. L’utilisation des Hormones en Elevage: Les Développements Zootechniques et les Préoccupations de Santé Publique. 2008. Available online: https://hal.archives-ouvertes.fr/hal-01173447 (accessed on 25 May 2021).
- Gotoh, N.; Moroda, K.; Watanabe, H.; Yoshinaga, K.; Tanaka, M.; Mizobe, H.; Ichioka, K.; Tokairin, S.; Wada, S. Metabolism of odd-numbered fatty acids and even-numbered fatty acids in mouse. J. Oleo Sci. 2008, 57, 293–299. [Google Scholar] [CrossRef] [Green Version]
- Collins, J.R.; Edwards, B.R.; Fredricks, H.F.; Van Mooy, B.A.S. LOBSTAHS: An Adduct-Based Lipidomics Strategy for Discovery and Identification of Oxidative Stress Biomarkers. Anal. Chem. 2016, 88, 7154–7162. [Google Scholar] [CrossRef]
- Koelmel, J.P.; Kroeger, N.M.; Ulmer, C.Z.; Bowden, J.A.; Patterson, R.E.; Cochran, J.A.; Beecher, C.W.W.; Garrett, T.J.; Yost, R.A. LipidMatch: An automated workflow for rule-based lipid identification using untargeted high-resolution tandem mass spectrometry data. BMC Bioinform. 2017, 18, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Huang, J.Z. Sparse principal component analysis via regularized low rank matrix approximation. J. Multivar. Anal. 2008, 99, 1015–1034. [Google Scholar] [CrossRef] [Green Version]
- Cao, K.-A.L.; Boitard, S.; Besse, P. Sparse PLS discriminant analysis: Biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinform. 2011, 12, 253. [Google Scholar] [CrossRef] [Green Version]
- Rinaudo, P.; Boudah, S.; Junot, C.; Thévenot, E.A. biosigner: A New Method for the Discovery of Significant Molecular Signatures from Omics Data. Front. Mol. Biosci. 2016, 3, 26. [Google Scholar] [CrossRef] [PubMed]


| Platform | Non-Targeted RPLC-HRMS [12] | Targeted Lipidyzer™ [33,34] | Targeted TG Platform [35] |
|---|---|---|---|
| Extraction type | Bligh and Dyer—like [12] | Two solvent addition/organic phase transfer cycles | Bligh and Dyer—[12] |
| Samples | D3, D9, D16, D18, D23 and D29 QC | D3, D18 and D23 QC Lipidyzer-specific QC and QC spike samples | D3, D16, D18, D23 and D29 QC |
| Serum volume | 30 µL | 30 µL | 10 µL, completed with 20 µL H2O |
| Solvents | Methanol (MeOH), Chloroform (CHCl3), Water (H2O) | MeOH, dichloromethane (DCM), H2O | MeOH, CHCl3, H2O |
| Centrifugation | Yes | Yes, two times | Yes |
| Internal standards | n = 7 In CHCl3, 0.5 mg·L−1 | Lipidyzer™ standard kit, n = 54 30 µL added at beginning (See Supplementary Materials) | n = 3 In CHCl3, 0.132 µmol·L−1 |
| Transfer | 200 µL organic phase | Multiple organic phases | 200 µL organic phase |
| Evaporation | Yes | Yes | Yes |
| Reconstitution solvent | Acetonitrile(AcN):Isopropanol(IPA):H2O (65:30:5, v:v:v) | DCM:MeOH (50:50, v:v), 10 mM Ammonium Acetate | AcN:IPA (50:50, v:v) |
| Reconstitution volume | 200 µL | 300 µL | 200 µL |
| Analysis Technique | LC-HRMS (full-scan + data dependent MS/MS) | DMS-MS/MS (direct introduction) | LC-MS/MS |
| Quantification | No | Yes | No |
| Targeted | No | Yes | Yes |
| Analytical system | LC: Thermo UltiMate® 3000 MS: Thermo Q-Exactive | Sciex QTRAP 5500, with SelexION differential mobility spectrometry (DMS) | LC: Waters Acquity UPLC MS: Waters Acquity-Synapt G2S Q-TOF |
| Column | Waters CSH C18 (100 × 2.1 mm i.d., 1.7 µm particle size) | None (direct introduction) | Waters BEH C18 (150 × 2.1 mm i.d. 1.7 µm particle size) |
| Mobile phase | A: ACN:H2O (60:40, v:v) B: IPA:ACN:H2O (88:10:2, v:v:v) Both: 10 mM ammonium acetate + 0.1% acetic acid | DCM:MeOH (50:50, v:v) 10 mM Ammonium Acetate | A: MeOH B: MeOH/IPA (50:50, v:v) Both: 2 mM ammonium acetate + 6 mM acetic acid |
| Ionisation | Polarity switching mode ESI− and ESI+ | Polarity switching mode ESI− and ESI+ | ESI+ |
| Data processing | MSConvert [36] XCMS [37], CAMERA Batch drift correction [38] Annotation: Lipidsearch (Thermo Fisher Scientific) after additional data dependent MS/MS—Top 15 (Full MS/dd-MS2-Top 15) acquisitions | Automated Lipidyzer™ framework | MassWolf XCMS [37] In-house R algorithm |
| Number of features/lipids in analysed samples | ESI−: 1612 features ESI+: 2914 features | 873 lipids * | 50 TG ** |
| Quality Assurance/Quality Control | Randomisation, QC (pooled samples), Internal standards, Extraction blanks | Randomisation, QC (pooled samples), Control plasma, Spiked samples, Internal standards, Extraction blanks | Cross checking of platform performance [35], calibration, QC (pooled samples), extraction blanks |
| Variable ID | VIPpred † | Annotation (LipidSearch) | MS2 Validation (LipidSearch) | p-Value D16 | p-Value D18 | p-Value D23 | p-Value D29 |
|---|---|---|---|---|---|---|---|
| ESI− | |||||||
| M791T491 | 1.81 | [PC(18:1_14:0) + CH3COO]− | ✓ | 0.117 | * 0.027 | 0.117 | * 0.034 |
| M805T538 | 1.98 | [PC(15:0_18:1) + CH3COO]− | ✓ | ** 0.009 | * 0.014 | * 0.028 | * 0.034 |
| M833T633 | 1.84 | [PC(17:0_18:1) + CH3COO]− | ✓ | * 0.028 | * 0.014 | 0.076 | * 0.034 |
| M715T534 | 1.92 | [PE(16:0_18:2)-H]− | ✓ | 0.117 | 0.221 | ** 0.009 | 0.480 |
| M717T611 | 2.18 | [PE(16:0_18:1)-H]− | ✓ | * 0.028 | 0.806 | ** 0.009 | 0.480 |
| M739T518 | 1.97 | [PE(16:0_20:4)-H]− | ✓ | * 0.047 | 0.086 | * 0.016 | 0.480 |
| M745T705 | 2.05 | [PE(18:0_18:1)-H]− | ✓ | 0.076 | 0.142 | * 0.047 | 0.289 |
| M753T566 | 2.02 | [PE(17:0_20:4)-H]− | ✓ | * 0.028 | * 0.014 | 0.076 | 0.480 |
| M765T524 | 2.17 | [PE(18:1_20:4)-H]− | ✓ | 0.076 | 0.142 | * 0.016 | 0.157 |
| M723T563 | 1.90 | [PE(16:0p_20:4)-H]− | ✓ | ** 0.009 | 0.221 | ** 0.009 | 0.077 |
| M751T659 | 1.84 | [PE(16:0p_22:4)-H]− | ✓ | ** 0.009 | 0.327 | * 0.028 | 0.157 |
| M829T472 | 1.80 | [PS(18:2_21:0)-H]− | ✓ | * 0.016 | * 0.050 | * 0.028 | * 0.034 |
| ESI+ | |||||||
| M777T719 | 1.95 | [PC(16:0_19:0) + H]+ | X | * 0.047 | * 0.027 | 0.175 | 0.289 |
| M755T566 | 1.81 | [PE(17:0_20:4) + H]+ | ✓ | ** 0.009 | * 0.014 | * 0.047 | 0.077 |
| M759T836 | 1.84 | [PE(20:0p_18:1) + H]+ | ✓ | * 0.028 | * 0.050 | * 0.047 | 0.077 |
| M865T1051 | 1.87 | [TG(16:0_17:0_18:1) + NH4]+ | ✓ | 0.117 | 0.086 | 0.076 | 0.157 |
| M879T1059 | 1.92 | [TG(18:0_16:0_18:1) + NH4]+ | ✓ | 0.076 | 0.142 | * 0.028 | 0.157 |
| M891T1051 | 1.84 | [TG(17:0_18:1_18:1) + NH4]+ | ✓ | 0.117 | 0.086 | * 0.047 | 0.157 |
| M893T1066 | 1.91 | [TG(18:0_17:0_18:1) + NH4]+ | ✓ | 0.076 | 0.086 | 0.076 | 0.289 |
| M898T1065 | 1.84 | [TG(18:0_17:0_18:1) + Na]+ | ✓ | 0.117 | 0.086 | 0.076 | 0.157 |
| M921T1080 | 2.35 | [TG(18:0_18:1_19:0) + NH4]+ | ✓ | 0.117 | 0.086 | * 0.047 | 0.157 |
| M926T1080 | 1.99 | [TG(18:0_18:1_19:0) + Na]+ | ✓ | * 0.047 | 0.086 | 0.076 | 0.157 |
| M919T1066 | 1.88 | [TG(19:1_18:0_18:1) + NH4]+ | ✓ | * 0.047 | 0.142 | * 0.016 | 0.077 |
| M924T1066 | 1.84 | [TG(19:0_18:1_18:1) + Na]+ | ✓ | 0.076 | 0.142 | * 0.047 | 0.157 |
| Lipid Class | p-Value D3 | p-Value D18 | p-Value D23 |
|---|---|---|---|
| CE | 0.55 | *0.03 | 0.10 |
| CER | 0.22 | 0.11 | 0.31 |
| DAG | 0.42 | 0.20 | ** 0.01 |
| DCER | 0.42 | 1.00 | 0.22 |
| FFA | 1.00 | 0.20 | 0.42 |
| HCER | 0.15 | * 0.03 | 0.69 |
| LCER | 0.69 | * 0.03 | ** 0.01 |
| LPC | 0.06 | 0.34 | 0.84 |
| LPE | 0.15 | 0.11 | 0.55 |
| PC | 0.69 | 0.06 | 0.06 |
| PE | 0.84 | * 0.03 | ** 0.01 |
| SM | 0.22 | * 0.03 | 1.00 |
| TG | 0.55 | 0.20 | 0.06 |
| TG_Rt | Corresponding Regioisomers with Estimated Proportions | p-Values | ||||
|---|---|---|---|---|---|---|
| D3 | D16 | D18 | D23 | D29 | ||
| TG(52:5)_553.44s | TG(rac-18:3/16:0/18:2)15% TG(rac-16:0/18:2/18:3)50% TG(rac-16:0/18:3/18:2)35% | 0.44 | 0.77 | 0.64 | * 0.03 | 0.06 |
| TG(54:6)_555.9s | TG(18:2/18:2/18:2) | 0.17 | * 0.05 | 0.39 | * 0.03 | 0.72 |
| TG(54:6)_566.5s | TG(rac-18:3/18:1/18:2)60% TG(rac-18:1/18:2/18:3)10% TG(rac-18:1/18:3/18:2)30% | 0.17 | 0.18 | 0.25 | *0.03 | 1.00 |
| TG(54:5)_685.8s | TG(rac-18:3/18:0/18:2)60% TG(rac-18:0/18:2/18:3)20% TG(rac-18:0/18:3/18:2)20% | 1.00 | 0.65 | 0.15 | * 0.05 | 0.51 |
| TG(54:7)_476.03s | TG(rac-18:2/18:2/18:3)60% TG(rac-18:2/18:3/18:2)40% | 0.65 | * 0.05 | 0.64 | * 0.03 | 1.00 |
| Non-Targeted RP LC-HRMS | Lipidyzer™ | TG Platform | ||||
|---|---|---|---|---|---|---|
| Class of the Relevant Lipids | Analysed and Annotated? | Variation (If Significant) | Analysed and Annotated? | Variation (If Significant) | Analysed and Annotated? | Variation (If Significant) |
| CE | Yes † | Yes | ↗D18 * ↗ D23 * | No | - | |
| CER | Yes † | Yes | ↗ D18 * | No | - | |
| DAG | Yes † | Yes | ↗ D18 * ↗ D23 * | No | - | |
| DCER | Yes † | Yes | ↗ D18 * | No | - | |
| FFA | Yes † | Yes | ↗ D18 * ↗ D23 * | No | - | |
| HCER | Yes † | Yes | ↘ D3 * ↗ D18 * | No | - | |
| LCER | No | Yes | ↗D18 * ↗ D23 * | No | - | |
| LPC | Yes † | Yes | - | No | - | |
| LPE | Yes † | Yes | ↗ D18 * | No | - | |
| PC | Yes | ↗ D16 *, ↗ D18 *, ↗ D23 *, ↗ D29 * | Yes | ↗ D18 * ↗ D23 * | No | - |
| PE | Yes | ↗ D16 *, ↗ D18 *, ↗ D23 * | Yes | ↗ D3 * ↗ D18 * ↗ D23 * | No | - |
| PS | Yes | ↗ D16 *, ↗ D18 *, ↗ D23 *, ↗ D29 * | No | - | No | - |
| SM | Yes † | Yes | ↗ D18 * | No | - | |
| TG | Yes | ↗ D16 *, ↗ D23 * | Yes | ↗ D18 * ↗ D23 * | Yes | ↘ D16 *, ↗ D23 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marchand, J.; Guitton, Y.; Martineau, E.; Royer, A.-L.; Balgoma, D.; Le Bizec, B.; Giraudeau, P.; Dervilly, G. Extending the Lipidome Coverage by Combining Different Mass Spectrometric Platforms: An Innovative Strategy to Answer Chemical Food Safety Issues. Foods 2021, 10, 1218. https://doi.org/10.3390/foods10061218
Marchand J, Guitton Y, Martineau E, Royer A-L, Balgoma D, Le Bizec B, Giraudeau P, Dervilly G. Extending the Lipidome Coverage by Combining Different Mass Spectrometric Platforms: An Innovative Strategy to Answer Chemical Food Safety Issues. Foods. 2021; 10(6):1218. https://doi.org/10.3390/foods10061218
Chicago/Turabian StyleMarchand, Jérémy, Yann Guitton, Estelle Martineau, Anne-Lise Royer, David Balgoma, Bruno Le Bizec, Patrick Giraudeau, and Gaud Dervilly. 2021. "Extending the Lipidome Coverage by Combining Different Mass Spectrometric Platforms: An Innovative Strategy to Answer Chemical Food Safety Issues" Foods 10, no. 6: 1218. https://doi.org/10.3390/foods10061218
APA StyleMarchand, J., Guitton, Y., Martineau, E., Royer, A. -L., Balgoma, D., Le Bizec, B., Giraudeau, P., & Dervilly, G. (2021). Extending the Lipidome Coverage by Combining Different Mass Spectrometric Platforms: An Innovative Strategy to Answer Chemical Food Safety Issues. Foods, 10(6), 1218. https://doi.org/10.3390/foods10061218