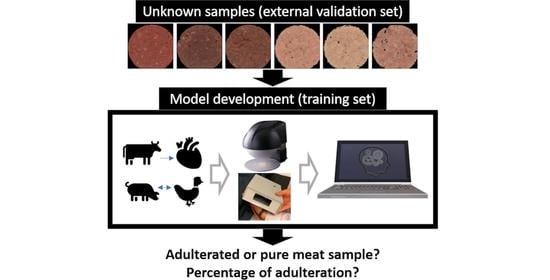
Detection of Meat Adulteration Using Spectroscopy-Based Sensors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Measurements Using Spectroscopy-Based Sensors and Data Acquisition
2.2.1. Vis and Fluo Data
2.2.2. MSI Data
2.3. Data Analysis
3. Results
3.1. Pork-Chicken Adulteration Scenario
3.2. Beef-Offal Adulteration Scenario
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- The EU Food Fraud Network and The System for Administrative Assistance & Food Fraud. European Commission, 2018. Available online: https://ec.europa.eu/food/sites/food/files/safety/docs/ff_ffn_annual-report_2018.pdf/ (accessed on 14 April 2021).
- Ellis, D.I.; Muhamadali, H.; Haughey, S.A.; Elliott, C.T.; Goodacre, R. Point-and-Shoot: Rapid Quantitative Detection Methods for on-Site Food Fraud Analysis—Moving out of the Laboratory and into the Food Supply Chain. Anal. Methods 2015, 7, 9401–9414. [Google Scholar] [CrossRef] [Green Version]
- Martuscelli, M.; Serio, A.; Capezio, O.; Mastrocola, D. Safety, Quality and Analytical Authentication of Ḥalāl Meat Products, with Particular Emphasis on Salami: A Review. Foods 2020, 9, 1111. [Google Scholar] [CrossRef] [PubMed]
- FAO. Meat Market Review—APRIL 2020. Available online: http://www.fao.org/3/ca8819en/CA8819EN.pdf (accessed on 13 April 2021).
- Mullen, A.M.; Álvarez, C.; Zeugolis, D.I.; Henchion, M.; O’Neill, E.; Drummond, L. Alternative Uses for Co-Products: Harnessing the Potential of Valuable Compounds from Meat Processing Chains. Meat Sci. 2017, 132, 90–98. [Google Scholar] [CrossRef] [PubMed]
- European Commission. The EU Food Fraud Network and the Administrative Assistance and Cooperation System: 2019 Annual Report. European Commission. Available online: https://ec.europa.eu/food/sites/food/files/safety/docs/ff_ffn_annual-report_2019.pdf/ (accessed on 14 April 2021).
- Montowska, M.; Pospiech, E. Authenticity Determination of Meat and Meat Products on the Protein and DNA Basis. Food Rev. Int. 2010, 27, 84–100. [Google Scholar] [CrossRef]
- Ballin, N.Z. Authentication of Meat and Meat Products. Meat Sci. 2010, 86, 577–587. [Google Scholar] [CrossRef]
- Candoğan, K.; Altuntas, E.G.; İğci, N. Authentication and Quality Assessment of Meat Products by Fourier-Transform Infrared (FTIR) Spectroscopy. Food Eng. Rev. 2020. [Google Scholar] [CrossRef]
- Alikord, M.; Momtaz, H.; Keramat, J.; Kadivar, M.; Rad, A.H. Species Identification and Animal Authentication in Meat Products: A Review. J. Food Meas. Charact. 2018, 12, 145–155. [Google Scholar] [CrossRef]
- Ballin, N.Z.; Vogensen, F.K.; Karlsson, A.H. Species Determination—Can We Detect and Quantify Meat Adulteration? Meat Sci. 2009, 83, 165–174. [Google Scholar] [CrossRef] [PubMed]
- European Parliament Resolution of 14 January 2014 on the Food Crisis, Fraud in the Food Chain and the Control Thereof (2013/2091(INI)). 2014. 9. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:52014IP0011(01)/ (accessed on 14 April 2021).
- Beganović, A.; Hawthorne, L.; Bach, K.; Huck, C. Critical Review on the Utilization of Handheld and Portable Raman Spectrometry in Meat Science. Foods 2019, 8, 49. [Google Scholar] [CrossRef] [Green Version]
- Porep, J.U.; Kammerer, D.R.; Carle, R. On-Line Application of near Infrared (NIR) Spectroscopy in Food Production. Trends Food Sci. Technol. 2015, 46, 211–230. [Google Scholar] [CrossRef]
- Kumar, Y.; Chandrakant Karne, S. Spectral Analysis: A Rapid Tool for Species Detection in Meat Products. Trends Food Sci. Technol. 2017, 62, 59–67. [Google Scholar] [CrossRef]
- Markiewicz-Keszycka, M.; Cama-Moncunill, R.; Pietat Casado-Gavalda, M.; Sullivan, C.; Cullen, P.J. Laser-Induced Breakdown Spectroscopy for Food Authentication. Curr. Opin. Food Sci. 2019, 28, 96–103. [Google Scholar] [CrossRef]
- Rady, A.; Adedeji, A. Application of Hyperspectral Imaging and Machine Learning Methods to Detect and Quantify Adulterants in Minced Meats. Food Anal Methods 2020, 13, 970–981. [Google Scholar] [CrossRef]
- Ropodi, A.I.; Panagou, E.Z.; Nychas, G.-J.E. Multispectral Imaging (MSI): A Promising Method for the Detection of Minced Beef Adulteration with Horsemeat. Food Control 2017, 73, 57–63. [Google Scholar] [CrossRef]
- Black, C.; Chevallier, O.P.; Cooper, K.M.; Haughey, S.A.; Balog, J.; Takats, Z.; Elliott, C.T.; Cavin, C. Rapid Detection and Specific Identification of Offals within Minced Beef Samples Utilising Ambient Mass Spectrometry. Sci. Rep. 2019, 9, 6295. [Google Scholar] [CrossRef] [Green Version]
- Rady, A.; Adedeji, A. Assessing Different Processed Meats for Adulterants Using Visible-near-Infrared Spectroscopy. Meat Sci. 2018, 136, 59–67. [Google Scholar] [CrossRef]
- Ropodi, A.I.; Panagou, E.Z.; Nychas, G.-J.E. Rapid Detection of Frozen-Then-Thawed Minced Beef Using Multispectral Imaging and Fourier Transform Infrared Spectroscopy. Meat Sci. 2018, 135, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wu, T.; Liu, Y.; Zou, J.; Huang, Y.; Babu, V.S.; Lin, L. Rapid Identification of Pork Adulterated in the Beef and Mutton by Infrared Spectroscopy. J. Spectrosc. 2018, 2018, 1–10. [Google Scholar] [CrossRef]
- Iammarino, M.; Marino, R.; Albenzio, M. How Meaty? Detection and Quantification of Adulterants, Foreign Proteins and Food Additives in Meat Products. Int. J. Food Sci. Technol. 2017, 52, 13. [Google Scholar] [CrossRef]
- Jiménez-Carvelo, A.M.; González-Casado, A.; Bagur-González, M.G.; Cuadros-Rodríguez, L. Alternative Data Mining/Machine Learning Methods for the Analytical Evaluation of Food Quality and Authenticity—A Review. Food Res. Int. 2019, 122, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Alamprese, C.; Amigo, J.M.; Casiraghi, E.; Engelsen, S.B. Identification and Quantification of Turkey Meat Adulteration in Fresh, Frozen-Thawed and Cooked Minced Beef by FT-NIR Spectroscopy and Chemometrics. Meat Sci. 2016, 121, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.-T.; Feng, Y.-Z.; Chen, W.; Jia, G.-F. Application of Invasive Weed Optimization and Least Square Support Vector Machine for Prediction of Beef Adulteration with Spoiled Beef Based on Visible Near-Infrared (Vis-NIR) Hyperspectral Imaging. Meat Sci. 2019, 151, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Tsakanikas, P.; Fengou, L.-C.; Manthou, E.; Lianou, A.; Panagou, E.Z.; Nychas, G.-J.E. A Unified Spectra Analysis Workflow for the Assessment of Microbial Contamination of Ready-to-Eat Green Salads: Comparative Study and Application of Non-Invasive Sensors. Comput. Electron. Agric. 2018, 155, 212–219. [Google Scholar] [CrossRef] [Green Version]
- Carstensen, J.M.; Hansen, J.F. Apparatus and a Method of Recording an Image of an Object. International Patent 17,198, 15 November 2003. [Google Scholar]
- Videometer A/S. Available online: http://www.videometer.com (accessed on 17 February 2021).
- Panagou, E.Z.; Papadopoulou, O.; Carstensen, J.M.; Nychas, G.-J.E. Potential of Multispectral Imaging Technology for Rapid and Non-Destructive Determination of the Microbiological Quality of Beef Filets during Aerobic Storage. Int. J. Food Microbiol. 2014, 174, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Barker, M.; Rayens, W. Partial Least Squares for Discrimination. J. Chemom. 2003, 17, 166–173. [Google Scholar] [CrossRef]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-Regression: A Basic Tool of Chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Luts, J.; Ojeda, F.; Van de Plas, R.; De Moor, B.; Van Huffel, S.; Suykens, J.A.K. A Tutorial on Support Vector Machine-Based Methods for Classification Problems in Chemometrics. Anal. Chim. Acta 2010, 665, 129–145. [Google Scholar] [CrossRef] [PubMed]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://Www.R-Project.Org/ (accessed on 14 April 2021).
- RStudio Team. RStudio: Integrated Development Environment for R; RStudio, PBC: Boston, MA, USA, 2020; Available online: http://Www.Rstudio.Com/ (accessed on 14 April 2021).
- Meyer, D.; Dimitriadou, E.; Hornik, K.; Weingessel, A.; Leisch, F. E1071: Misc Functions of the Department of Statistics, Probability Theory Group (Formerly: E1071), TU Wien, R Package Version 1.7-4. Available online: https://cran.r-project.org/web/packages/e1071/e1071.pdf/ (accessed on 14 April 2021).
- Mevik, B.-H.; Wehrens, R.; Liland, K.H. Pls: Partial Least Squares and Principal Component Regression, R Package Version 2.7-3. Available online: https://cran.r-project.org/web/packages/pls/pls.pdf/ (accessed on 14 April 2021).
- Kuhn, M. Caret: Classification and Regression Training, R Package Version 6.0-86. Available online: https://cran.r-project.org/web/packages/caret/caret.pdf/ (accessed on 14 April 2021).
- Landis, J.R.; Koch, G.G. The Measurement of Observer Agreement for Categorical Data. Biometrics 1977, 33, 159. [Google Scholar] [CrossRef] [Green Version]
- Warrens, M.J. Five Ways to Look at Cohen’s Kappa. J. Psychol. Psychother. 2015, 5, 1. [Google Scholar] [CrossRef] [Green Version]
- Nolasco-Perez, I.M.; Rocco, L.A.C.M.; Cruz-Tirado, J.P.; Pollonio, M.A.R.; Barbon, S.; Barbon, A.P.A.C.; Barbin, D.F. Comparison of Rapid Techniques for Classification of Ground Meat. Biosyst. Eng. 2019, 183, 151–159. [Google Scholar] [CrossRef]
- Fengou, L.-C.; Tsakanikas, P.; Nychas, G.-J.E. Rapid Detection of Minced Pork and Chicken Adulteration in Fresh, Stored and Cooked Ground Meat. Food Control 2021, 125, 108002. [Google Scholar] [CrossRef]
- Velioglu, H.M.; Sezer, B.; Bilge, G.; Baytur, S.E.; Boyaci, I.H. Identification of Offal Adulteration in Beef by Laser Induced Breakdown Spectroscopy (LIBS). Meat Sci. 2018, 138, 28–33. [Google Scholar] [CrossRef]
- Casado-Gavalda, M.P.; Dixit, Y.; Geulen, D.; Cama-Moncunill, R.; Cama-Moncunill, X.; Markiewicz-Keszycka, M.; Cullen, P.J.; Sullivan, C. Quantification of Copper Content with Laser Induced Breakdown Spectroscopy as a Potential Indicator of Offal Adulteration in Beef. Talanta 2017, 169, 123–129. [Google Scholar] [CrossRef]
- Hu, Y.; Zou, L.; Huang, X.; Lu, X. Detection and Quantification of Offal Content in Ground Beef Meat Using Vibrational Spectroscopic-Based Chemometric Analysis. Sci. Rep. 2017, 7, 15162. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Downey, G.; O’Donnell, C.P. Dispersive Raman Spectroscopy and Multivariate Data Analysis To Detect Offal Adulteration of Thawed Beefburgers. J. Agric. Food Chem. 2015, 63, 1433–1441. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; O’Donnell, C.P.; Downey, G. Detection of Offal Adulteration in Beefburgers Using near Infrared Reflectance Spectroscopy and Multivariate Modelling. J. Infrared Spectrosc. 2013, 21, 237–248. [Google Scholar] [CrossRef]
- Morsy, N.; Sun, D.-W. Robust Linear and Non-Linear Models of NIR Spectroscopy for Detection and Quantification of Adulterants in Fresh and Frozen-Thawed Minced Beef. Meat Sci. 2013, 11, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Al-Sarayreh, M.; Reis, M.M.; Yan, W.Q.; Klette, R. Detection of Red-Meat Adulteration by Deep Spectral–Spatial Features in Hyperspectral Images. J. Imaging 2018, 4, 63. [Google Scholar] [CrossRef] [Green Version]
- Kamruzzaman, M.; Makino, Y.; Oshita, S. Rapid and Non-Destructive Detection of Chicken Adulteration in Minced Beef Using Visible near-Infrared Hyperspectral Imaging and Machine Learning. J. Food Eng. 2016, 170, 8–15. [Google Scholar] [CrossRef]
- Zheng, X.; Li, Y.; Wei, W.; Peng, Y. Detection of Adulteration with Duck Meat in Minced Lamb Meat by Using Visible Near-Infrared Hyperspectral Imaging. Meat Sci. 2019, 149, 55–62. [Google Scholar] [CrossRef]
- Ropodi, A.I.; Pavlidis, D.E.; Mohareb, F.; Panagou, E.Z.; Nychas, G.-J.E. Multispectral Image Analysis Approach to Detect Adulteration of Beef and Pork in Raw Meats. Food Res. Int. 2015, 67, 12–18. [Google Scholar] [CrossRef]
- Aït-Kaddour, A.; Loudiyi, M.; Ferlay, A.; Gruffat, D. Performance of Fluorescence Spectroscopy for Beef Meat Authentication: Effect of Excitation Mode and Discriminant Algorithms. Meat Sci. 2018, 137, 58–66. [Google Scholar] [CrossRef]
- Hassoun, A.; Sahar, A.; Lakhal, L.; Aït-Kaddour, A. Fluorescence Spectroscopy as a Rapid and Non-Destructive Method for Monitoring Quality and Authenticity of Fish and Meat Products: Impact of Different Preservation Conditions. LWT 2019, 103, 279–292. [Google Scholar] [CrossRef]
- McGrath, T.F.; Haughey, S.A.; Patterson, J.; Fauhl-Hassek, C.; Donarski, J.; Alewijn, M.; van Ruth, S.; Elliott, C.T. What Are the Scientific Challenges in Moving from Targeted to Non-Targeted Methods for Food Fraud Testing and How Can They Be Addressed?—Spectroscopy Case Study. Trends Food Sci. Technol. 2018, 76, 38–55. [Google Scholar] [CrossRef]
- Tsakanikas, P.; Karnavas, A.; Panagou, E.Z.; Nychas, G.J. A Machine Learning Workflow for Raw Food Spectroscopic Classification in a Future Industry. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]


| True Class | ||||||
|---|---|---|---|---|---|---|
| Sensors | 0% | 25% | 50% | 75% | 100% | |
| Vis fresh samples | Specificity (%) | 100.00 | 100.00 | 58.33 | 91.67 | 95.83 |
| Recall (%) | 33.33 | 33.33 | 100.00 | 66.67 | 50.00 | |
| Precision (%) | 100.00 | 100.00 | 37.50 | 66.67 | 75.00 | |
| F1-score | 0.50 | 0.50 | 0.54 | 0.67 | 0.60 | |
| Accuracy (%) | 56.67 | |||||
| Kappa | 0.46 | |||||
| MSI fresh samples | Specificity (%) | 100.00 | 100.00 | 100.00 | 87.50 | 100.00 |
| Recall (%) | 100.00 | 100.00 | 100.00 | 100.00 | 50.00 | |
| Precision (%) | 100.00 | 100.00 | 100.00 | 66.67 | 100.00 | |
| F1-score | 1.00 | 1.00 | 1.00 | 0.80 | 0.67 | |
| Accuracy (%) | 90.00 | |||||
| Kappa | 0.87 | |||||
| MSI frozen-thawed samples | Specificity (%) | 100.00 | 100.00 | 100.00 | 95.83 | 87.50 |
| Recall (%) | 100.00 | 100.00 | 83.33 | 50.00 | 100.00 | |
| Precision (%) | 100.00 | 100.00 | 100.00 | 75.00 | 66.67 | |
| F1-score | 1.00 | 1.00 | 0.91 | 0.60 | 0.80 | |
| Accuracy (%) | 86.67 | |||||
| Kappa | 0.83 | |||||
| True Class | ||||
|---|---|---|---|---|
| Sensors | 0% | A | 100% | |
| Vis fresh samples | Specificity (%) | 100.00 | 83.33 | 100.00 |
| Recall (%) | 100.00 | 100.00 | 66.67 | |
| Precision (%) | 100.00 | 90.00 | 100.00 | |
| F1-score | 1.00 | 0.95 | 0.80 | |
| Accuracy (%) | 93.33 | |||
| Kappa | 0.87 | |||
| MSI fresh samples | Specificity (%) | 100.00 | 100.00 | 100.00 |
| Recall (%) | 100.00 | 100.00 | 100.00 | |
| Precision (%) | 100.00 | 100.00 | 100.00 | |
| F1-score | 100.00 | 100.00 | 100.00 | |
| Accuracy (%) | 100.00 | |||
| Kappa | 1.00 | |||
| MSI frozen-thawed samples | Specificity (%) | 100.00 | 100.00 | 91.67 |
| Recall (%) | 100.00 | 88.89 | 100.00 | |
| Precision (%) | 100.00 | 100.00 | 75.00 | |
| F1-score | 1.00 | 0.94 | 0.86 | |
| Accuracy (%) | 93.33 | |||
| Kappa | 0.89 | |||
| True Class | ||||||
|---|---|---|---|---|---|---|
| Sensors | 0% | 25% | 50% | 75% | 100% | |
| Vis fresh samples | Specificity (%) | 100.00 | 70.83 | 100.00 | 100.00 | 100.00 |
| Recall (%) | 100.00 | 100.00 | 0.00 | 83.33 | 100.00 | |
| Precision (%) | 100.00 | 46.15 | 1 NaN | 100.00 | 100.00 | |
| F1-score | 1.00 | 0.63 | 1 NaN | 0.91 | 1.00 | |
| Accuracy (%) | 76.67 | |||||
| Kappa | 0.62 | |||||
| MSI fresh samples | Specificity (%) | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
| Recall (%) | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | |
| Precision (%) | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | |
| F1-score | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| Accuracy (%) | 100.00 | |||||
| Kappa | 1.00 | |||||
| MSI frozen-thawed samples | Specificity (%) | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
| Recall (%) | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | |
| Precision (%) | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | |
| F1-score | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| Accuracy (%) | 100.00 | |||||
| Kappa | 1.00 | |||||
| True Class | ||||
|---|---|---|---|---|
| Sensors | 0% | A | 100% | |
| Vis fresh samples | Specificity (%) | 100.00 | 91.67 | 100.00 |
| Recall (%) | 83.33 | 100.00 | 100.00 | |
| Precision (%) | 100.00 | 94.74 | 100.00 | |
| F1-score | 0.91 | 0.97 | 1.00 | |
| Accuracy (%) | 96.67 | |||
| Kappa | 0.94 | |||
| MSI fresh samples | Specificity (%) | 100.00 | 83.33 | 100.00 |
| Recall (%) | 100.00 | 100.00 | 66.67 | |
| Precision (%) | 100.00 | 90.00 | 100.00 | |
| F1-score | 1.00 | 0.95 | 0.80 | |
| Accuracy (%) | 93.33 | |||
| Kappa | 0.87 | |||
| MSI frozen-thawed samples | Specificity (%) | 100.00 | 100.00 | 100.00 |
| Recall (%) | 100.00 | 100.00 | 100.00 | |
| Precision (%) | 100.00 | 100.00 | 100.00 | |
| F1-score | 1.00 | 1.00 | 1.00 | |
| Accuracy (%) | 100.00 | |||
| Kappa | 1.00 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fengou, L.-C.; Lianou, A.; Tsakanikas, P.; Mohareb, F.; Nychas, G.-J.E. Detection of Meat Adulteration Using Spectroscopy-Based Sensors. Foods 2021, 10, 861. https://doi.org/10.3390/foods10040861
Fengou L-C, Lianou A, Tsakanikas P, Mohareb F, Nychas G-JE. Detection of Meat Adulteration Using Spectroscopy-Based Sensors. Foods. 2021; 10(4):861. https://doi.org/10.3390/foods10040861
Chicago/Turabian StyleFengou, Lemonia-Christina, Alexandra Lianou, Panagiοtis Tsakanikas, Fady Mohareb, and George-John E. Nychas. 2021. "Detection of Meat Adulteration Using Spectroscopy-Based Sensors" Foods 10, no. 4: 861. https://doi.org/10.3390/foods10040861
APA StyleFengou, L. -C., Lianou, A., Tsakanikas, P., Mohareb, F., & Nychas, G. -J. E. (2021). Detection of Meat Adulteration Using Spectroscopy-Based Sensors. Foods, 10(4), 861. https://doi.org/10.3390/foods10040861