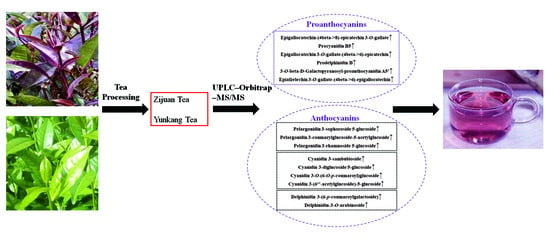
Analysis of Differentiated Chemical Components between Zijuan Purple Tea and Yunkang Green Tea by UHPLC-Orbitrap-MS/MS Combined with Chemometrics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Tea Samples
2.2. Sample Preparation
2.3. Ultra-High Performance Liquid Chromatography Combined with Orbitrap Mass Spectrometry (UHPLC-Orbitrap-MS/MS) Analysis
2.4. Catechins and Caffeine Analysis by HPLC
2.5. Total Content of Anthocyanins by Acid-Alcohol Method
2.6. Data Analysis for Untargeted Metabolomics
3. Results
3.1. Significant Differentiation of Zijuan Purple Tea (ZJT) and Yunkang Green Tea (YKT) Metabolite Profiling by UHPLC-Orbitrap-MS/MS
3.2. Characteristic Metabolites of ZJT
3.2.1. Flavan-3-Ols
3.2.2. Proanthocyanins
3.2.3. Flavonol and Flavone Glycosides
3.2.4. Amino Acids and Phenolic Acids
3.2.5. Alkaloids
3.3. Metabolic Pathway Analysis of ZJT
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Meng, X.-H.; Li, N.; Zhu, H.-T.; Wang, D.; Yang, C.-R.; Zhang, Y.-J. Plant Resources, Chemical Constituents, and Bioactivities of Tea Plants from the Genus Camellia Section Thea. J. Agric. Food Chem. 2018, 67, 5318–5349. [Google Scholar] [CrossRef]
- Fraser, K.; Harrison, S.J.; Lane, G.A.; Otter, D.E.; Hemar, Y.; Quek, S.-Y.; Rasmussen, S. Analysis of Low Molecular Weight Metabolites in Tea Using Mass Spectrometry-Based Analytical Methods. Crit. Rev. Food Sci. Nutr. 2014, 54, 924–937. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Hu, O.; Fu, H.; Ouyang, L.; Gong, X.; Meng, P.; Wang, Z.; Dai, M.; Guo, X.; Wang, Y. UPLC–Q-TOF/MS-based untargeted metabolomics coupled with chemometrics approach for Tieguanyin tea with seasonal and year variations. Food Chem. 2019, 283, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Du, F.; Zhou, L.; Jiao, Y.; Bai, S.; Wang, L.; Ma, J.; Fu, X. Ingredients in Zijuan Pu’er Tea Extract Alleviate β-Amyloid Peptide Toxicity in a Caenorhabditis elegans Model of Alzheimer’s Disease Likely through DAF-16. Molecules 2019, 24, 729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, Y.; Xia, L.; Li, Y.; Liang, M. A new tea tree cultivar ‘Zijuan’. Acta Hortic. Sin. 2008, 35, 934. [Google Scholar] [CrossRef]
- Jiang, L.; Shen, X.; Shoji, T.; Kanda, T.; Zhou, J.; Zhao, L. Characterization and Activity of Anthocyanins in Zijuan Tea (Camellia sinensis var. kitamura). J. Agric. Food Chem. 2013, 61, 3306–3310. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.R.; Bao, Y.X.; Huang, M. The botanical and quality characteristics of the tea cultivar “zi-Juan” in Yunnan province. J. Tea 2009, 35, 17–18. [Google Scholar] [CrossRef]
- Gao, X.; Ho, C.-T.; Li, X.; Lin, X.; Zhang, Y.; Chen, Z.; Li, B. Phytochemicals, Anti-Inflammatory, Antiproliferative, and Methylglyoxal Trapping Properties of Zijuan Tea. J. Food Sci. 2018, 83, 517–524. [Google Scholar] [CrossRef]
- Wang, Q.-P.; Peng, C.-X.; Gao, B.; Gong, J.-S. Influence of large molecular polymeric pigments isolated from fermented Zijuan tea on the activity of key enzymes involved in lipid metabolism in rat. Exp. Gerontol. 2012, 47, 672–679. [Google Scholar] [CrossRef]
- Gong, J.; Zhang, Q.; Peng, C.; Fan, J.; Dong, W. Curie-point pyrolysis–gas chromatography–mass spectroscopic analysis of theabrownins from fermented Zijuan tea. J. Anal. Appl. Pyrolysis 2012, 97, 171–180. [Google Scholar] [CrossRef]
- Jing, J.; Shi, Y.; Zhang, Q.; Wang, J.; Ruan, J. Prediction of Chinese green tea ranking by metabolite profiling using ultra-performance liquid chromatography–quadrupole time-of-flight mass spectrometry (UPLC–Q-TOF/MS). Food Chem. 2017, 221, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Li, N.; Zhao, M.; Yu, W.; Wu, J.-L. Metabolomic profiling delineate taste qualities of tea leaf pubescence. Food Res. Int. 2017, 94, 36–44. [Google Scholar] [CrossRef]
- Xu, J.; Hu, F.-L.; Wang, W.; Wan, X.-C.; Bao, G.-H. Investigation on biochemical compositional changes during the microbial fermentation process of Fu brick tea by LC–MS based metabolomics. Food Chem. 2015, 186, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Dai, W.; Lu, M.; Lv, H.; Guo, L.; Zhang, Y.; Zhu, Y.; Peng, Q.; Lin, Z. Study of the dynamic changes in the non-volatile chemical constituents of black tea during fermentation processing by a non-targeted metabolomics approach. Food Res. Int. 2016, 79, 106–113. [Google Scholar] [CrossRef]
- Dai, W.; Xie, D.; Lu, M.; Li, P.; Lv, H.; Yang, C.; Peng, Q.; Zhu, Y.; Guo, L.; Zhang, Y.; et al. Characterization of white tea metabolome: Comparison against green and black tea by a nontargeted metabolomics approach. Food Res. Int. 2017, 96, 40–45. [Google Scholar] [CrossRef]
- Chen, S.; Liu, H.; Zhao, X.; Li, X.; Shan, W.; Wang, X.; Wang, S.; Yu, W.; Yang, Z.; Yu, X. Non-targeted metabolomics analysis reveals dynamic changes of volatile and non-volatile metabolites during oolong tea manufacture. Food Res. Int. 2020, 128, 108778. [Google Scholar] [CrossRef]
- Long, P.; Wen, M.; Granato, D.; Zhou, J.; Wu, Y.; Hou, Y.; Zhang, L. Untargeted and targeted metabolomics reveal the chemical characteristic of pu-erh tea (Camellia assamica) during pile-fermentation. Food Chem. 2020, 311, 125895. [Google Scholar] [CrossRef]
- Xin, Z.; Ma, S.; Ren, D.; Liu, W.; Han, B.; Zhang, Y.; Xiao, J.; Yi, L.; Deng, B. UPLC–Orbitrap–MS/MS combined with chemometrics establishes variations in chemical components in green tea from Yunnan and Hunan origins. Food Chem. 2018, 266, 534–544. [Google Scholar] [CrossRef]
- Zhang, Q.; Hu, J.; Liu, M.; Shi, Y.; De Vos, R.C.H.; Ruan, J. Stimulated biosynthesis of delphinidin-related anthocyanins in tea shoots reducing the quality of green tea in summer. J. Sci. Food Agric. 2019, 100, 1505–1514. [Google Scholar] [CrossRef]
- Zeng, C.; Lin, H.; Liu, Z.; Liu, Z. Metabolomics analysis of Camellia sinensis with respect to harvesting time. Food Res. Int. 2020, 128, 108814. [Google Scholar] [CrossRef]
- Ghaste, M.; Mistrik, R.; Shulaev, V. Applications of Fourier Transform Ion Cyclotron Resonance (FT-ICR) and Orbitrap Based High Resolution Mass Spectrometry in Metabolomics and Lipidomics. Int. J. Mol. Sci. 2016, 17, 816. [Google Scholar] [CrossRef]
- Wang, W.; Fu, X.-W.; Dai, X.-L.; Hua, F.; Chu, G.-X.; Chu, M.-J.; Hu, F.-L.; Ling, T.-J.; Gao, L.-P.; Xie, Z.-W.; et al. Novel acetylcholinesterase inhibitors from Zijuan tea and biosynthetic pathway of caffeoylated catechin in tea plant. Food Chem. 2017, 237, 1172–1178. [Google Scholar] [CrossRef]
- Available online: http://xcmsonline.scripps.edu/ (accessed on 16 December 2020).
- Teng, Y.; Li, D.; Guruvaiah, P.; Xu, N.; Xie, Z. Dietary Supplement of Large Yellow Tea Ameliorates Metabolic Syndrome and Attenuates Hepatic Steatosis in db/db Mice. Nutrients 2018, 10, 75. [Google Scholar] [CrossRef] [Green Version]
- Patti, G.J.; Tautenhahn, R.; Siuzdak, G. Meta-analysis of untargeted metabolomic data from multiple profiling experiments. Nat. Protoc. 2012, 7, 508–516. [Google Scholar] [CrossRef] [Green Version]
- Zeng, C.; Lin, H.; Liu, Z.; Liu, Z. Analysis of Young Shoots of ‘Anji Baicha’ (Camellia sinensis) at Three Developmental Stages Using Nontargeted LC-MS-Based Metabolomics. J. Food Sci. 2019, 84, 1746–1757. [Google Scholar] [CrossRef]
- De Vos, R.C.H.; Moco, S.; Lommen, A.; Keurentjes, J.J.B.; Bino, R.J.; Hall, R.D. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2007, 2, 778–791. [Google Scholar] [CrossRef]
- Qin, D.; Wang, Q.; Li, H.; Jiang, X.; Fang, K.; Wang, Q.; Li, B.; Pan, C.; Wu, H. Identification of key metabolites based on non-targeted metabolomics and chemometrics analyses provides insights into bitterness in Kucha [Camellia kucha (Chang et Wang) Chang]. Food Res. Int. 2020, 138, 109789. [Google Scholar] [CrossRef]
- Ji, H.-G.; Lee, Y.-R.; Lee, M.-S.; Hwang, K.H.; Park, C.Y.; Kim, E.-H.; Park, J.S.; Hong, Y.-S. Diverse Metabolite Variations in Tea (Camellia sinensis L.) Leaves Grown Under Various Shade Conditions Revisited: A Metabolomics Study. J. Agric. Food Chem. 2018, 66, 1889–1897. [Google Scholar] [CrossRef] [PubMed]
- Available online: http://www.hmdb.ca/ (accessed on 12 February 2021).
- Available online: http://www.ncbi.nlm.nih.gov/pccompound/ (accessed on 20 January 2021).
- Available online: http://metlin.scripps.edu (accessed on 20 January 2021).
- Yue, W.; Sun, W.; Rao, R.S.P.; Ye, N.; Yang, Z.; Chen, M. Non-targeted metabolomics reveals distinct chemical compositions among different grades of Bai Mudan white tea. Food Chem. 2019, 277, 289–297. [Google Scholar] [CrossRef]
- Available online: http://www.genome.jp/kegg/pathway.html (accessed on 20 January 2021).
- Yue, Y.; Chu, G.-X.; Liu, X.-S.; Tang, X.; Wang, W.; Liu, G.-J.; Yang, T.; Ling, T.-J.; Wang, X.-G.; Zhang, Z.-Z.; et al. TMDB: A literature-curated database for small molecular compounds found from tea. BMC Plant Biol. 2014, 14, 243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarachiwin, L.; Ute, K.; Kobayashi, A.; Fukusaki, E. 1H NMR Based Metabolic Profiling in the Evaluation of Japanese Green Tea Quality. J. Agric. Food Chem. 2007, 55, 9330–9336. [Google Scholar] [CrossRef]
- Dixon, R.A.; Xie, D.; Sharma, S.B. Proanthocyanidins—A final frontier in flavonoid research? New Phytol. 2005, 165, 9–28. [Google Scholar] [CrossRef] [Green Version]
- Khoo, H.E.; Azlan, A.; Tang, S.T.; Lim, S.M. Anthocyanidins and anthocyanins: Colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 2017, 61, 1361779. [Google Scholar] [CrossRef] [Green Version]
- Cheng, S.; Fu, X.; Wang, X.; Liao, Y.; Zeng, L.; Dong, F.; Yang, Z. Studies on the Biochemical Formation Pathway of the Amino Acidl-Theanine in Tea (Camellia sinensis) and Other Plants. J. Agric. Food Chem. 2017, 65, 7210–7216. [Google Scholar] [CrossRef]
- Chen, S.; Li, M.; Zheng, G.; Wang, T.; Lin, J.; Wang, S.; Wang, X.; Chao, Q.; Cao, S.; Yang, Z.; et al. Metabolite Profiling of 14 Wuyi Rock Tea Cultivars Using UPLC-QTOF MS and UPLC-QqQ MS Combined with Chemometrics. Molecules 2018, 23, 104. [Google Scholar] [CrossRef] [Green Version]
- Willstätter, R.; Everest, A.E. Untersuchungen über die Anthocyane. I. Über den Farbstoff der Kornblume. Eur. J. Org. Chem. 1913, 401, 189–232. [Google Scholar] [CrossRef] [Green Version]
- Yu, Z.; Yang, Z. Understanding different regulatory mechanisms of proteinaceous and non-proteinaceous amino acid formation in tea (Camellia sinensis) provides new insights into the safe and effective alteration of tea flavor and function. Crit. Rev. Food Sci. Nutr. 2020, 60, 844–858. [Google Scholar] [CrossRef]
- Wei, K.; Wang, L.-Y.; Zhou, J.; He, W.; Zeng, J.-M.; Jiang, Y.-W.; Cheng, H. Comparison of catechins and purine alkaloids in albino and normal green tea cultivars (Camellia sinensis L.) by HPLC. Food Chem. 2012, 130, 720–724. [Google Scholar] [CrossRef]
- Yan, X.; Wang, Z.; Mei, Y.; Wang, L.; Wang, X.; Xu, Q.; Peng, S.; Zhou, Y.; Wei, C. Isolation, Diversity, and Growth-Promoting Activities of Endophytic Bacteria From Tea Cultivars of Zijuan and Yunkang-10. Front. Microbiol. 2018, 9, 1848. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Tian, H.-L.; Wu, J.-H.; Cao, R.-R.; Wang, R.-X.; Qi, X.-H.; Xu, Q.; Chen, X.-H. Relationship between gene expression and the accumulation of catechin during spring and autumn in tea plants (Camellia sinensis L.). Hortic. Res. 2015, 2, 15023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, H.P.; Yang, T.; Liang, M.Z.; Wang, L.B.; Lin, Z. Study of EGCG3″Me content in Zijuan tea. Mod. Food Sci. Technol. 2014, 30, 286–289. [Google Scholar] [CrossRef]
- Kurita, I.; Maeda-Yamamoto, M.; Tachibana, H.; Kamei, M. Antihypertensive Effect of Benifuuki Tea ContainingO-Methylated EGCG. J. Agric. Food Chem. 2010, 58, 1903–1908. [Google Scholar] [CrossRef] [PubMed]
- Oritani, Y.; Setoguchi, Y.; Ito, R.; Maruki-Uchida, H.; Ichiyanagi, T.; Ito, T. Comparison of (−)-Epigallocatechin-3-O-gallate (EGCG) and O-Methyl EGCG Bioavailability in Rats. Biol. Pharm. Bull. 2013, 36, 1577–1582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, J.; Zou, Z.; Zhang, X.; Zhou, L.; Wang, Y.; Fang, W.; Zhu, X. Metabolic analyses reveal different mechanisms of leaf color change in two purple-leaf tea plant (Camellia sinensis L.) cultivars. Hortic. Res. 2018, 5, 7. [Google Scholar] [CrossRef] [Green Version]
- Pollastri, S.; Tattini, M. Flavonols: Old compounds for old roles. Ann. Bot. 2011, 108, 1225–1233. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Pan, D.; Liang, M.; Abubakar, Y.S.; Li, J.; Lin, J.; Chen, S.; Chen, W. Regulation of Anthocyanin Biosynthesis in Purple Leaves of Zijuan Tea (Camellia sinensis var. kitamura). Int. J. Mol. Sci. 2017, 18, 833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Liu, M.; Ruan, J. Metabolomics analysis reveals the metabolic and functional roles of flavonoids in light-sensitive tea leaves. BMC Plant Biol. 2017, 17, 64. [Google Scholar] [CrossRef] [Green Version]
- Mei, Y.; Xie, H.; Liu, S.; Zhu, J.; Zhao, S.; Wei, C. Metabolites and Transcriptional Profiling Analysis Reveal the Molecular Mechanisms of the Anthocyanin Metabolism in the “Zijuan” Tea Plant (Camellia sinensis var. assamica). J. Agric. Food Chem. 2021, 69, 414–427. [Google Scholar] [CrossRef]





| Compound Name | RT (min) | Detected Mass | Theoretical Mass | Adduct | Formula | MS/MS Fragment | VIP | p-Value | |Log2 FC| | FC |
|---|---|---|---|---|---|---|---|---|---|---|
| Flavan-3-ols | ||||||||||
| Epiafzelechin 3-O-gallate | 4.30 | 426.0945 | 426.0951 | [M-H2O-H]- | C22H18O9 | 169, 125 | 1.38 | 2.00 × 10−9 | 1.30 | 2.46 |
| Epicatechin 3-O-gallate | 4.49 | 442.0925 | 442.0905 | [M-H2O-H]- | C22H18O10 | 123, 139, 153 | 1.37 | 3.00 × 10−8 | 1.97 | 3.91 |
| Catechin * | 4.60 | 290.0785 | 290.0790 | [M+H]+ | C15H14O6 | 95,97,109,123,125 | 1.23 | 2.00 × 10−4 | 0.83 | 1.78 |
| Epigallocatechin gallate * | 5.02 | 458.0831 | 458.0849 | [M+H]+ | C22H18O11 | 139, 151, 289 | 1.45 | 4.00 × 10−3 | 1.31 | 0.40 |
| Epigallocatechin 3-(3”-methylgallate) | 5.96 | 472.1006 | 472.1006 | [M-H]- | C23H20O11 | 139, 140, 167 | 1.38 | 4.00 × 10−9 | 6.06 | 66.66 |
| Epicatechin 3-O-(3-O-methylgallate) | 7.32 | 456.1055 | 456.1056 | [M-H]- | C23H20O10 | 109, 124, 168, 183, 289 | 1.38 | 2.00 × 10−8 | 7.61 | 195.68 |
| Epicatechin * | 7.34 | 290.0793 | 290.0790 | [M-H]- | C15H14O6 | 123, 139, 147, 207 | 1.37 | 2.00 × 10−8 | 3.57 | 11.87 |
| Epigallocatechin 3-p-coumaroate | 8.12 | 452.1109 | 452.1107 | [M-H]- | C24H20O9 | 196, 255 | 1.37 | 5.00 × 10−8 | 1.60 | 3.03 |
| Proanthocyanins | ||||||||||
| Epigallocatechin-(4beta->8)-epicatechin 3-O-gallate | 4.27 | 746.1481 | 746.1483 | [M+H]+ | C37H30O17 | 305, 443, 579 | 1.36 | 3.00 × 10−7 | 1.73 | 3.32 |
| Procyanidin B5 | 5.25 | 578.1430 | 578.1424 | [M-H2O-H]- | C30H26O12 | 289, 469 | 1.37 | 8.00 × 10−8 | 3.05 | 8.29 |
| 3-O-beta-D-Galactopyranosylproanthocyanidin A5′ | 5.50 | 738.1771 | 738.1796 | [M+Na]+ | C36H34O17 | 195, 455, 723 | 1.31 | 8.00 × 10−6 | 9.93 | 976.56 |
| Prodelphinidin B | 6.01 | 610.1335 | 610.1323 | [M+H]+ | C30H26O14 | 309, 757 | 1.38 | 2.00 × 10−9 | 2.46 | 5.49 |
| Epigallocatechin 3-O-gallate-(4beta->6)-epicatechin 3-O-gallate | 6.67 | 898.1590 | 898.1593 | [M+H]+ | C44H34O21 | 433, 741 | 1.32 | 5.00 × 10−6 | 1.39 | 2.62 |
| Epiafzelechin 3-O-gallate-(4beta->6)-epigallocatechin 3-O-gallate | 7.41 | 882.1608 | 882.1643 | [M+H]+ | C44H34O20 | 271, 441, 591 | 1.25 | 8.00 × 10−5 | 1.53 | 2.89 |
| Flavonol and flavone glycosides | ||||||||||
| Myricetin | 3.65 | 318.0383 | 318.0376 | [M-H2O-H]- | C15H8O7 | 125, 169, 241 | 1.39 | 9.00 × 10−10 | 4.31 | 19.85 |
| Vitexin 2″-O-rhamnoside-4‴-acetate | 3.77 | 620.1727 | 620.1741 | [M-H2O-H]- | C29H32O15 | 339, 449, 493 | 1.16 | 9.00 × 10−4 | 0.69 | 1.61 |
| Kaempferol 3-O-arabinoside | 4.05 | 418.0891 | 418.0900 | [M+H]+ | C20H18O10 | 211, 401 | 1.37 | 7.00 × 10−8 | 5.07 | 33.57 |
| Quercetin 3-galloylglucosyl-arabinofuranoside | 4.32 | 748.1520 | 748.1487 | [M-H]- | C33H32O20 | 229, 285 | 1.38 | 8.00 × 10−9 | 2.46 | 5.50 |
| Myricetin 3-rhamnoside-3′-glucoside | 4.95 | 626.1495 | 626.1483 | [M-H]- | C27H30O17 | 178, 316 | 1.05 | 5.00 × 10−3 | 0.72 | 1.64 |
| Isovitexin 2″-O-rhamnoside | 4.95 | 578.1621 | 578.1636 | [M+H]+ | C27H30O14 | 247, 291, 409, 427 | 1.24 | 1.00 × 10−4 | 2.67 | 6.36 |
| Kaempferol 3-glucoside | 5.27 | 448.1007 | 448.1006 | [M-H2O-H]- | C21H20O11 | 145, 301, 377 | 1.29 | 2.00 × 10−5 | 1.34 | 2.52 |
| Kaempferol 7,4′-dirhamnoside | 5.50 | 578.1629 | 578.1636 | [M-H]- | C27H30O14 | 245, 289 | 1.26 | 6.00 × 10−5 | 2.75 | 6.72 |
| Pelargonidin 3-rhamnoside 5-glucoside | 5.50 | 579.1722 | 579.1714 | [M-H]- | C27H31O14 | 154, 245, 289, 469 | 1.25 | 1.00 × 10−4 | 2.79 | 6.90 |
| Cyanidin 3-sambubioside | 5.63 | 581.1510 | 581.1506 | [M-H]- | C26H29O15 | 419, 435, 449, 458 | 1.28 | 3.00 × 10−5 | 0.99 | 1.99 |
| Quercetin 3-rutinoside-4′-glucoside | 5.90 | 772.2045 | 772.2062 | [M-H]- | C33H40O21 | 151, 255, 271, 301, 300 | 1.31 | 7.00 × 10−6 | 9.73 | 849.64 |
| Cyanidin 3-diglucoside 5-glucoside | 5.90 | 773.2145 | 773.2140 | [M-H]- | C33H41O21 | 277, 513 | 1.30 | 1.00 × 10−5 | 11.33 | 2579.02 |
| Kaempferol 3-(2″-hydroxypropionylglucoside)-4′-glucoside | 5.94 | 682.1734 | 682.1745 | [M-H2O-H]- | C30H34O18 | 213, 249, 327 | 1.36 | 3.00 × 10−7 | 4.87 | 29.33 |
| Delphinidin 3-(6-p-coumaroylgalactoside) | 6.01 | 611.1379 | 611.1401 | [M+H]+ | C30H27O14 | 138, 331, 409 | 1.38 | 2.00 × 10−9 | 2.65 | 6.29 |
| Quercetagetin 7-methylether 6-glucoside | 6.04 | 494.1060 | 494.1060 | [M-H]- | C22H22O13 | 163, 319, 337 | 1.30 | 1.00 × 10−5 | 2.04 | 4.10 |
| Quercetin 3-arabinoside | 6.11 | 434.0853 | 434.0849 | [M-H]- | C20H18O11 | 343, 313 | 1.36 | 1.00 × 10−7 | 2.15 | 4.43 |
| Delphinidin-3-O-arabinoside | 6.12 | 435.0933 | 435.0927 | [M-H]- | C20H19O11 | 165, 289, 341 | 1.26 | 8.00 × 10−5 | 2.11 | 4.33 |
| Quercetin 3-galactoside | 6.14 | 464.0944 | 464.0955 | [M+H]+ | C21H20O12 | 138, 303 | 1.08 | 3.00 × 10−3 | 0.91 | 1.88 |
| Cyanidin 3-(6″-acetylglucoside)-5-glucoside | 6.31 | 652.1641 | 652.1639 | [M-H2O-H]- | C29H32O17 | 207, 315, 515 | 1.08 | 3.00 × 10−3 | 0.63 | 1.55 |
| Pelargonidin 3-sophoroside 5-glucoside | 6.32 | 757.2184 | 757.2191 | [M+H]+ | C33H41O20 | 287, 433, 595 | 1.39 | 5.00 × 10−10 | 5.98 | 63.09 |
| Cyanidin 3-O-(6-O-p-coumaroyl)glucoside | 6.46 | 594.1373 | 594.1373 | [M+H]+ | C30H26O13 | 166, 273, 424, 442, 527 | 1.39 | 2.00 × 10−10 | 2.17 | 4.49 |
| Kaempferol 3-(6″-ferulylglucoside) | 6.51 | 624.1464 | 624.1479 | [M+H]+ | C31H28O14 | 317, 165, 203 | 1.33 | 2.00 × 10−6 | 8.49 | 360.45 |
| Pelargonidin 3-coumarylglucoside-5-acetylglucoside | 6.52 | 782.2061 | 782.2058 | [M-H2O-H]- | C38H38O18 | 457, 461, 337 | 1.31 | 1.00 × 10−5 | 3.73 | 13.26 |
| Kaempferol 3-O-[2″-(4‴--acetyl-rhamnosyl)-6″-glucosyl] glucoside | 6.53 | 798.2216 | 798.2219 | [M+H]+ | C35H42O21 | 519, 524 | 1.36 | 2.00 × 10−7 | 4.01 | 16.15 |
| Kaempferol 3-glucoside-7-xyloside | 6.83 | 580.1452 | 580.1428 | [M+H]+ | C26H28O15 | 271, 396 | 1.33 | 2.00 × 10−6 | 3.93 | 15.27 |
| Kaempferol 3-(4″-acetyl-6″-p-coumarylglucoside) | 8.14 | 636.1456 | 636.1479 | [M-H2O-H]- | C29H32O16 | 313, 465, 483 | 1.36 | 2.00 × 10−7 | 3.16 | 8.93 |
| Quercetin 3,3′-dimethyl ether 4′-glucoside | 8.58 | 510.1369 | 510.1373 | [M-H2O-H]- | C23H26O13 | 285, 442, 599 | 1.18 | 5.00 × 10−4 | 1.04 | 2.05 |
| Kaempferol | 8.90 | 286.0465 | 286.0477 | [M+H]+ | C15H10O6 | 121, 165, 241 | 1.55 | 4.00 × 10−3 | 1.80 | 0.29 |
| Phenolic acids | ||||||||||
| Phenol | 0.74 | 94.0414 | 94.0419 | [M+H]+ | C6H6O | 62, 73, 82 | 1.23 | 4.00 × 10−3 | 1.24 | 0.42 |
| 5-O-Caffeoylquinic acid | 1.51 | 354.0955 | 354.0951 | [M-H]+ | C16H18O9 | 103, 175 | 1.36 | 2.00 × 10−7 | 2.33 | 5.02 |
| Chlorogenic acid | 2.42 | 354.0933 | 354.0951 | [M+Na]+ | C16H18O9 | 93, 135, 173, 191 | 1.47 | 4.00 × 10−3 | 0.71 | 0.61 |
| Salicylic acid | 4.24 | 138.0309 | 138.0317 | [M+H]+ | C7H6O3 | 56, 65, 116, 139, 140 | 1.52 | 4.00 × 10−3 | 1.44 | 0.37 |
| 4-Hydroxybenzoic acid | 5.02 | 138.0312 | 138.0317 | [M+H]+ | C7H6O3 | 56, 111, 116, 139, 140 | 1.40 | 4.00 × 10−3 | 0.69 | 0.62 |
| 3-O-Methylgallate | 7.35 | 184.0376 | 184.0372 | [M-H]- | C8H8O5 | 124, 139, 168 | 1.33 | 3.00 × 10−6 | 0.97 | 1.97 |
| 3-O-p-Coumaroylquinic acid | 7.54 | 338.1000 | 338.1002 | [M-H2O-H]- | C16H18O8 | 93, 119, 173, 191 | 1.15 | 1.00 × 10−3 | 1.31 | 2.48 |
| Shikimic acid | 9.49 | 174.0521 | 174.0528 | [M-H2O-H]- | C7H10O5 | 61, 67, 93, 173 | 1.33 | 4.00 × 10−3 | 0.68 | 0.62 |
| Amino acids | ||||||||||
| Lysine | 0.63 | 146.1050 | 146.1055 | [M+H]+ | C6H14N2O2 | 72, 84, 128, 130 | 1.36 | 4.00 × 10−3 | 1.01 | 0.50 |
| Alanine | 0.72 | 89.0472 | 89.0477 | [M+H]+ | C3H7NO2 | 68, 77 | 1.27 | 4.00 × 10−3 | 0.88 | 0.54 |
| Aspartic acid | 0.74 | 133.0371 | 133.0375 | [M+H]+ | C4H7NO4 | 74, 88, 102, 116 | 1.15 | 4.00 × 10−3 | 1.84 | 0.28 |
| Methionine | 1.04 | 149.0503 | 149.0510 | [M+H]+ | C5H11NO2S | 102, 131 | 1.23 | 4.00 × 10−3 | 1.37 | 0.39 |
| Valine | 1.05 | 117.0785 | 117.0790 | [M+H]+ | C5H11NO2 | 58, 59, 118, 119 | 1.32 | 4.00 × 10−3 | 1.04 | 0.49 |
| Isoleucine | 1.92 | 131.0939 | 131.0946 | [M+H]+ | C6H13NO2 | 69, 72, 86, 90 | 1.39 | 4.00 × 10−3 | 0.89 | 0.54 |
| Tyrosine | 2.76 | 181.1882 | 181.1885 | [M-H]- | C9H11NO3 | 91, 119, 123, 136, 165 | 1.37 | 7.00 × 10−8 | 3.57 | 11.84 |
| Cysteine | 5.63 | 121.1586 | 121.1582 | [M-H]- | C3H7NO2S | 74, 100, 98 | 1.22 | 2.00 × 10−4 | 0.90 | 1.87 |
| Alkaloids | ||||||||||
| Adenine | 0.63 | 135.0556 | 135.0545 | [M-H]- | C5H5N5 | 119 | 1.28 | 4.00 × 10−3 | 0.84 | 0.56 |
| 2-Acetylpyrazine | 1.04 | 122.0475 | 122.0480 | [M+H]+ | C6H6N2O | 80, 95, 96, 123 | 1.28 | 4.00 × 10−3 | 1.91 | 0.27 |
| 3-Methylxanthine | 1.15 | 166.0487 | 166.0491 | [M+H]+ | C6H6N4O2 | 120 | 1.28 | 4.00 × 10−5 | 3.17 | 9.01 |
| Ellagic acid | 3.86 | 302.0048 | 302.0063 | [M+H]+ | C14H6O8 | 57, 275, 285, 303 | 1.20 | 4.00 × 10−3 | 1.71 | 0.31 |
| 2-Acetylpyrrole | 4.27 | 109.0526 | 109.0528 | [M+Hac-H]- | C6H7NO | 67, 83 | 1.29 | 4.00 × 10−3 | 1.42 | 0.37 |
| Theophylline | 6.27 | 180.0641 | 180.0647 | [M-H2O+H]- | C7H8N4O2 | 135, 146, 161, 164 | 1.50 | 4.00 × 10−3 | 0.62 | 0.65 |
| Aquifoliunine EII | 6.32 | 721.2591 | 721.2582 | [M+K]+ | C34H43NO16 | 231, 273, 411 | 1.39 | 2.00 × 10−10 | 4.82 | 28.31 |
| Caffeine * | 9.54 | 194.0800 | 194.0804 | [M-H2O-H]- | C8H10N4O2 | 85, 93 | 1.30 | 4.00 × 10−3 | 0.59 | 0.66 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Shen, Y.; Ling, T.; Ho, C.-T.; Li, D.; Guo, H.; Xie, Z. Analysis of Differentiated Chemical Components between Zijuan Purple Tea and Yunkang Green Tea by UHPLC-Orbitrap-MS/MS Combined with Chemometrics. Foods 2021, 10, 1070. https://doi.org/10.3390/foods10051070
Li M, Shen Y, Ling T, Ho C-T, Li D, Guo H, Xie Z. Analysis of Differentiated Chemical Components between Zijuan Purple Tea and Yunkang Green Tea by UHPLC-Orbitrap-MS/MS Combined with Chemometrics. Foods. 2021; 10(5):1070. https://doi.org/10.3390/foods10051070
Chicago/Turabian StyleLi, Mengwan, Ying Shen, Tiejun Ling, Chi-Tang Ho, Daxiang Li, Huimin Guo, and Zhongwen Xie. 2021. "Analysis of Differentiated Chemical Components between Zijuan Purple Tea and Yunkang Green Tea by UHPLC-Orbitrap-MS/MS Combined with Chemometrics" Foods 10, no. 5: 1070. https://doi.org/10.3390/foods10051070
APA StyleLi, M., Shen, Y., Ling, T., Ho, C. -T., Li, D., Guo, H., & Xie, Z. (2021). Analysis of Differentiated Chemical Components between Zijuan Purple Tea and Yunkang Green Tea by UHPLC-Orbitrap-MS/MS Combined with Chemometrics. Foods, 10(5), 1070. https://doi.org/10.3390/foods10051070