Building of an Internal Transcribed Spacer (ITS) Gene Dataset to Support the Italian Health Service in Mushroom Identification
Abstract
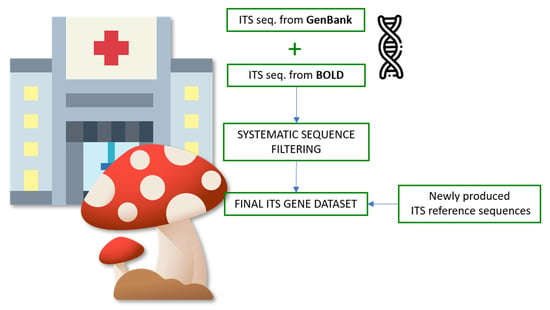
:1. Introduction
2. Materials and Methods
2.1. Target Species: Selection Criteria
2.2. ITS Sequences Retrieval from Public Genetic Databases
2.3. Systematic Double-Step ITS Sequences Filtering and Intra-Species Divergence Estimation
2.3.1. First Step: Sequence Quality and Technical Check
2.3.2. Second Step: Phylogenetic Analysis
2.3.3. Intra-Species Divergence Estimation
2.4. Samples Collection, Morphological Identification, and Production of ITS Internal Reference Sequences
2.4.1. Mushroom Samples Collection and Morphological Identification
2.4.2. Total DNA Extraction and Evaluation
2.4.3. ITS Region Amplification, Sequencing, and Sequence Editing
2.5. Building of the Final ITS Gene Dataset
2.5.1. Taxonomic Coverage
2.5.2. Evaluation of ITS Region Efficiency in Species Identification
2.6. ITS Gene Dataset Validation
3. Results and Discussion
3.1. Target Species Selection
3.2. ITS Sequence Retrieval from Public Databases
3.3. Systematic Double-Step ITS Sequences Filtering, Intra-Species Divergence Estimation, and Evaluation of ITS Region Efficiency
3.3.1. First Step: Sequences Quality and Technical Check
3.3.2. Second Step: Phylogenetic Analysis
3.3.3. Intra-Species Divergence Estimation
3.4. Samples Collection, Molecular Analysis, and Production of ITS Internal Reference Sequences
3.4.1. Mushroom Sample Collection
3.4.2. DNA Extraction, ITS Region Amplification, and ITS Reference Sequences Production
3.5. Building of the Final ITS Gene Dataset
3.5.1. Taxonomic Coverage Implementation of the ITS Gene Dataset
3.5.2. Evaluation of ITS Region Efficiency in Species Identification
3.6. Validation of the Final ITS Gene Dataset
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Blackwell, M. The Fungi: 1, 2, 3… 5.1 million species? Am. J. Bot. 2011, 98, 426–438. [Google Scholar] [CrossRef]
- Hibbett, D.; Abarenkov, K.; Kõljalg, U.; Öpik, M.; Chai, B.; Cole, J.; Wang, Q.; Crous, P.; Robert, V.; Helgason, T.; et al. Sequence-based classification and identification of Fungi. Mycologia 2016, 108, 1049–1068. [Google Scholar] [PubMed]
- Kalač, P. A review of chemical composition and nutritional value of wild-growing and cultivated mushrooms. J. Sci. Food Agric. 2013, 93, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Boa, E.R. Wild Edible Fungi: A Global Overview of Their Use and Importance to People; FAO: Rome, Italy, 2004. [Google Scholar]
- Kotowski, M. Differences between European regulations on wild mushroom commerce and actual trends in wild mushroom picking. Slov. Národopis 2016, 64, 169–178. [Google Scholar]
- Peintner, U.; Schwarz, S.; Mešić, A.; Moreau, P.A.; Moreno, G.; Saviuc, P. Mycophilic or mycophobic? Legislation and guidelines on wild mushroom commerce reveal different consumption behaviour in European countries. PLoS ONE 2013, 8, e63926. [Google Scholar] [CrossRef] [Green Version]
- Feeney, M.J.; Miller, A.M.; Roupas, P. Mushrooms—Biologically distinct and nutritionally unique: Exploring a “third food kingdom”. Nutr. Today 2014, 49, 301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giusti, A.; Ricci, E.; Gasperetti, L.; Galgani, M.; Polidori, L.; Verdigi, F.; Narducci, R.; Armani, A. Molecular identification of mushroom species in Italy: An ongoing project aimed at reinforcing the control measures of an increasingly appreciated sustainable food. Sustainability 2021, 13, 238. [Google Scholar] [CrossRef]
- Global Edible Mushroom Market Research Report. 2020. Available online: https://www.themarketreports.com/report/global-edible-mushroom-market-research-report (accessed on 11 April 2021).
- Sitta, N.; Floriani, M. Nationalization and globalization trends in the wild mushroom commerce of Italy with emphasis on porcini (Boletus edulis and allied species). Econ. Bot. 2008, 62, 307–322. [Google Scholar] [CrossRef]
- Govorushko, S.; Rezaee, R.; Dumanov, J.; Tsatsakis, A. Poisoning associated with the use of mushrooms: A review of the global pattern and main characteristics. Food Chem. Toxicol. 2019, 128, 267–279. [Google Scholar] [CrossRef] [PubMed]
- White, J.; Weinstein, S.A.; De Haro, L.; Bédry, R.; Schaper, A.; Rumack, B.H.; Zilker, T. Mushroom poisoning: A proposed new clinical classification. Toxicon 2019, 157, 53–65. [Google Scholar] [CrossRef]
- Erden, A.; Esmeray, K.; Karagöz, H.; Karahan, S.; Gümüşçü, H.H.; Başak, M.; Çetinkaya, A.; Avcı, D.; Poyrazoğlu, O.K. Acute liver failure caused by mushroom poisoning: A case report and review of the literature. Int. Med. Case Rep. 2013, 6, 85. [Google Scholar]
- Gawlikowski, T.; Romek, M.; Satora, L. Edible mushroom-related poisoning: A study on circumstances of mushroom collection, transport, and storage. Hum. Exp. Toxicol. 2015, 34, 718–724. [Google Scholar] [CrossRef]
- Nieminen, P.; Mustonen, A.M. Toxic potential of traditionally consumed mushroom species-a controversial continuum with many unanswered questions. Toxins 2020, 12, 639. [Google Scholar] [CrossRef]
- Bever, C.S.; Adams, C.A.; Hnasko, R.M.; Cheng, L.W.; Stanker, L.H. Lateral flow immunoassay (LFIA) for the detection of lethal amatoxins from mushrooms. PLoS ONE 2020, 15, e0231781. [Google Scholar] [CrossRef]
- Sitta, N.; Angelini, C.; Balma, M.; Berma, C.; Bertocchi, C.; Suriano, E. I funghi che causano intossicazioni in Italia. In Proceedings of the VI Mycotoxicology Conference, Perugia, Italy, 12–14 March 2018; pp. 23–24. [Google Scholar]
- Assisi, F.; Davanzo, F.; Bissoli, M.; Dimasi, V.; Ferruzzi, M.; Georgatos, J.; Rebutti, I.; Travaglia, A.; Severgnini, P.; Sesana, F.; et al. Epidemiologia e clinica delle intossicazioni da funghi in Italia: Valutazione retrospettiva di 20 anni (1998–2017) del Centro Antiveleni di Milano. Boll. Epidemiol. Naz. 2019. Available online: https://www.epicentro.iss.it/ben/2019/aprile/epidemiologia-intossicazione-funghi (accessed on 24 May 2021).
- Law of 23 August 1993 No. 352. Norme Quadro in Matera di Raccolta e Commercializzazione dei Funghi Epigei Freschi e Conservati. G.U. No. 215 of 13 September 1993. Available online: http://www.edizionieuropee.it/LAW/HTML/11/zn2_06_075.html (accessed on 11 April 2021).
- Presidential Decree of 14 July 1995 No. 376. Regolamento Concernente la Disciplina della Raccolta e della Commercializzazione dei Funghi Epigei Freschi e Conservati; G.U. No. 212 of 11 September 1995. Available online: http://www.salute.gov.it/imgs/C_17_normativa_1788_allegato.pdf (accessed on 11 April 2021).
- Raja, H.A.; Miller, A.N.; Pearce, C.J.; Oberlies, N.H. Fungal identification using molecular tools: A primer for the natural products research community. J. Nat. Prod. 2017, 80, 756–770. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W. Finding needles in haystacks: Linking scientific names, reference specimens and molecular data for Fungi. Database 2014, 109, 6241–6246. [Google Scholar] [CrossRef] [PubMed]
- Slepecky, R.A.; Starmer, W.T. Phenotypic plasticity in fungi: A review with observations on Aureobasidium pullulans. Mycologia 2009, 101, 823–832. [Google Scholar] [CrossRef] [PubMed]
- Epis, S.; Matinato, C.; Gentili, G.; Varotto, F.; Bandi, C.; Sassera, D. Molecular detection of poisonous mushrooms in different matrices. Mycologia 2010, 102, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Raja, H.A.; Baker, T.R.; Little, J.G.; Oberlies, N.H. DNA barcoding for identification of consumer-relevant mushrooms: A partial solution for product certification? Food Chem. 2017, 214, 383–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalho, L.M.; Carvalho, F.; de Lourdes Bastos, M.; Baptista, P.; Moreira, N.; Monforte, A.R.; da Silva Ferreira, A.C.; de Pinho, P.G. Non-targeted and targeted analysis of wild toxic and edible mushrooms using gas chromatography–ion trap mass spectrometry. Talanta 2014, 118, 292–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugawara, R.; Yamada, S.; Tu, Z.; Sugawara, A.; Suzuki, K.; Hoshiba, T.; Eisaka, S.; Yamaguchi, A. Rapid and reliable species identification of wild mushrooms by matrix assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS). Anal. Chim. Acta 2016, 934, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Badotti, F.; de Oliveira, F.S.; Garcia, C.F.; Vaz, A.B.M.; Fonseca, P.L.C.; Nahum, L.A.; Oliveira, G.; Góes-Neto, A. Effectiveness of ITS and sub-regions as DNA barcode markers for the identification of Basidiomycota (Fungi). BMC Microbiol. 2017, 17, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W. Fungal Barcoding Consortium. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Creedy, T.J.; Norman, H.; Tang, C.Q.; Qing Chin, K.; Andujar, C.; Arribas, P.; O’Connor, R.S.; Carvell, C.; Notton, D.G.; Vogler, A.P. A validated workflow for rapid taxonomic assignment and monitoring of a national fauna of bees (Apiformes) using high throughput DNA barcoding. Mol. Ecol. Resour. 2020, 20, 40–53. [Google Scholar] [CrossRef]
- Conde-Sousa, E.; Pinto, N.; Amorim, A. Reference DNA databases for forensic species identification: Auditing algorithms. Forensic Sci. Int. Genet. Suppl. Ser. 2019, 7, 564–566. [Google Scholar] [CrossRef]
- Chen, Q.; Zobel, J.; Verspoor, K. Duplicates, redundancies and inconsistencies in the primary nucleotide databases: A descriptive study. Database 2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giusti, A.; Guarducci, M.; Stern, N.; Davidovich, N.; Golani, D.; Armani, A. The importance of distinguishing pufferfish species (Lagocephalus spp.) in the Mediterranean Sea for ensuring public health: Evaluation of the genetic databases reliability in supporting species identification. Fish. Res. 2019, 210, 14–21. [Google Scholar] [CrossRef]
- Hofstetter, V.; Buyck, B.; Eyssartier, G.; Schnee, S.; Gindro, K. The unbearable lightness of sequenced-based identification. Fungal Divers. 2019, 96, 243–284. [Google Scholar] [CrossRef] [Green Version]
- Meiklejohn, K.A.; Damaso, N.; Robertson, J.M. Assessment of BOLD and GenBank–Their accuracy and reliability for the identification of biological materials. PLoS ONE 2019, 14, e0217084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pentinsaari, M.; Ratnasingham, S.; Miller, S.E.; Hebert, P.D. BOLD and GenBank revisited–Do identification errors arise in the lab or in the sequence libraries? PLoS ONE 2020, 15, e0231814. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.H.; Hyde, K.D.; Pawłowska, J.; Ryberg, M.; Tedersoo, L.; Aas, A.B.; Abarenkov, K. Improving ITS sequence data for identification of plant pathogenic fungi. Fungal Divers. 2014, 67, 11–19. [Google Scholar] [CrossRef]
- Lucas, S.G. Proper syntax when using aff. and cf. in taxonomic statements. J. Vertebr. Paleontol. 1986, 6, 202. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Hillis, D.M.; Bull, J.J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
- Armani, A.; Castigliego, L.; Tinacci, L.; Gandini, G.; Gianfaldoni, D.; Guidi, A. A rapid PCR–RFLP method for the identification of Lophius species. Eur. Food Res. Technol. 2012, 235, 253–263. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.J.W.T.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Hibbett, D.S. Major events in the evolution of the Fungi. Princet. Guide Evol. 2014, 152, 158. [Google Scholar]
- Loizides, M.; Bellanger, J.M.; Assyov, B.; Moreau, P.A.; Richard, F. Present status and future of boletoid fungi (Boletaceae) on the island of Cyprus: Cryptic and threatened diversity unravelled by ten-year study. Fungal Ecol. 2019, 41, 65–81. [Google Scholar] [CrossRef]
- Page, R.D. BioNames: Linking taxonomy, texts, and trees. PeerJ 2013, 1, e190. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, R.H.; Ryberg, M.; Kristiansson, E.; Abarenkov, K.; Larsson, K.H.; Kõljalg, U. Taxonomic reliability of DNA sequences in public sequence databases: A fungal perspective. PLoS ONE 2006, 1, e59. [Google Scholar] [CrossRef]
- Hughes, K.W.; Petersen, R.H.; Lickey, E.B. Using heterozygosity to estimate a percentage DNA sequence similarity for environmental species’ delimitation across basidiomycete fungi. New Phytol. 2009, 182, 795–798. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Kristiansson, E.; Ryberg, M.; Hallenberg, N.; Larsson, K.H. Intraspecific ITS variability in the kingdom Fungi as expressed in the international sequence databases and its implications for molecular species identification. Evol. Bioinform. 2008, 4, S653. [Google Scholar] [CrossRef] [PubMed]
- Amend, A.S.; Seifert, K.A.; Samson, R.; Bruns, T.D. Indoor fungal composition is geographically patterned and more diverse in temperate zones than in the tropics. Proc. Natl. Acad. Sci. USA 2010, 107, 13748–13753. [Google Scholar] [CrossRef] [Green Version]
- Gazis, R.; Rehner, S.; Chaverri, P. Species delimitation in fungal endophyte diversity studies and its implications in ecological and biogeographic inferences. Mol. Ecol. 2011, 20, 3001–3013. [Google Scholar] [CrossRef]
- Porras-Alfaro, A.; Liu, K.L.; Kuske, C.R.; Xie, G. From genus to phylum: Large-subunit and internal transcribed spacer rRNA operon regions show similar classification accuracies influenced by database composition. Appl. Environ. Microbiol. 2014, 80, 829–840. [Google Scholar] [CrossRef] [Green Version]
- Hollingsworth, M.L.; Clark, A.; Forrest, L.L.; Richardson, J.; Pennington, R.T.; Long, D.G.; Cowan, R.; Chase, M.W.; Gaudeul, M.; Hollingsworth, P.M. Selecting barcoding loci for plants: Evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Mol. Ecol. Resour. 2009, 9, 439–457. [Google Scholar] [CrossRef] [PubMed]
- Lücking, R.; Hawksworth, D.L. Formal description of sequence-based voucherless Fungi: Promises and pitfalls, and how to resolve them. IMA Fungus 2018, 9, 143–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, J.W.; Jacobson, D.J.; Kroken, S.; Kasuga, T.; Geiser, D.M.; Hibbett, D.S.; Fisher, M.C. Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 2000, 31, 21–32. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Aydin, M.; Bansal, G.; Kaya, E.; Singh, R. Determination of amatoxin concentration in heat-treated samples of Amanita phalloides by high-performance liquid chromatography: A forensic approach. J. Forensic Leg. Med. 2021, 78, 102111. [Google Scholar] [CrossRef] [PubMed]
- Schenk-Jaeger, K.M.; Rauber-Lüthy, C.; Bodmer, M.; Kupferschmidt, H.; Kullak-Ublick, G.A.; Ceschi, A. Mushroom poisoning: A study on circumstances of exposure and patterns of toxicity. Eur. J. Intern. Med. 2012, 23, e85–e91. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, M.; Carron, P.N.; Yersin, B.; Trueb, L. Mushroom poisoning: A retrospective study concerning 11-years of admissions in a Swiss Emergency Department. Intern. Emerg. Med. 2018, 13, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Wennig, R.; Eyer, F.; Schaper, A.; Zilker, T.; Andresen-Streichert, H. Mushroom poisoning. Dtsch. Arztebl. Int. 2020, 117, 701. [Google Scholar] [CrossRef]
- Luque, G.M.; Donlan, C.J. The characterization of seafood mislabeling: A global meta-analysis. Biol. Conserv. 2019, 236, 556–570. [Google Scholar] [CrossRef]
- Jensen-Vargas, E.; Marizzi, C. DNA Barcoding for identification of consumer-relevant fungi sold in New York: A powerful tool for citizen scientists? Foods 2018, 7, 87. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Mo, M.; Yang, L.; Mi, F.; Cao, Y.; Liu, C.; Tang, X.; Wang, P.; Xu, J. Exploring the species diversity of edible mushrooms in Yunnan, Southwestern China, by DNA barcoding. J. Fungi 2021, 7, 310. [Google Scholar] [CrossRef] [PubMed]
- Regulation (EU) 2017/625 of the European Parliament and of the Council of 15 March 2017 on official controls and other official activities performed to ensure the application of food and feed law, rules on animal health and welfare, plant health and plant protection products, amending Regulations (EC) No 999/2001, (EC) No 396/2005, (EC) No 1069/2009, (EC) No 1107/2009, (EU) No 1151/2012, (EU) No 652/2014, (EU) 2016/429 and (EU) 2016/2031 of the European Parliament and of the Council, Council Regulations (EC) No 1/2005 and (EC) No 1099/2009 and Council Directives 98/58/EC, 1999/74/EC, 2007/43/EC, 2008/119/EC and 2008/120/EC, and repealing Regulations (EC) No 854/2004 and (EC) No 882/2004 of the European Parliament and of the Council, Council Directives 89/608/EEC, 89/662/EEC, 90/425/EEC, 91/496/EEC, 96/23/EC, 96/93/EC and 97/78/EC and Council Decision 92/438/EEC (Official Controls Regulation). OJ L 95, 7.4. 2017, pp. 1–142. Available online: https://www.fsai.ie/uploadedFiles/Legislation/Food_Legisation_Links/Official_Control_Of_Foodstuffs/Consol_Reg2017_625.pdf (accessed on 24 May 2021).




| Cases (n) | Species/Genus | Edibility |
|---|---|---|
| 153 | Entoloma sinuatum | T |
| 135 | Omphalotus olearius | T |
| 22 | Clitocybe nebularis | T |
| 13 | Russula spp. | * |
| 10 | Boletus satanas (Rubroboletus satanas) | T |
| 9 | Agaricus xanthodermus | T |
| 9 | Macrolepiota venenata | T |
| 7 | Amanita phalloides | M |
| 7 | Macrolepiota rachodes (Chlorophyllum rhacodes) | SNE |
| 7 | Amanita muscaria/A. aureola (A. muscaria) | T |
| 6 | Armillara mellea | E |
| 5 | Inocybe spp. | * |
| 4 | Lepiota spp. | * |
| 3 | Amanita pantherina | T |
| 3 | Boletus luridus (Suillellus luridus) | E |
| 3 | Clitocybe spp. | * |
| 3 | Clitocybe rivulosa | T |
| 3 | Entoloma spp. | * |
| 3 | Lepiota josserandii/L. subincarnata (L. subincarnata) | M |
| 3 | Macrolepiota spp. | * |
| 3 | Psathyrella candolleana | T |
| 2 | Amanita ovoidea | T |
| 2 | Armillaria tabescens (Desarmillaria tabescens) | E |
| 2 | Lepiota cristata | T |
| 2 | Panaeolus spp. | * |
| 2 | Ramaria spp. | * |
| 2 | Tricholoma saponaceum | T |
| 1 | Agaricus spp. | * |
| 1 | Agaricus preclaresquamosus (Agaricus moelleri) | T |
| 1 | Amanita spp. | * |
| 1 | Amanita proxima | T |
| 1 | Amanita verna | M |
| 1 | Boletus lupinus (Rubroboletus lupinus) | T |
| 1 | Boletus pulchrotinctus (Suillellus pulchrotinctus) | T |
| 1 | Cantharellus cornucopioides | E |
| 1 | Clitocybe dealbata | T |
| 1 | Entoloma lividoalbum | SNE |
| 1 | Entoloma rhodopolium | SNE |
| 1 | Hygrophoropsis aurantiaca | SNE |
| 1 | Hypholoma fasciculare | T |
| 1 | Inocybe rimosa | T |
| 1 | Lactarius zonarius | T |
| 1 | Lepista nuda | E |
| 1 | Ramaria flavescens | E |
| 1 | Ramaria formosa | T |
| 1 | Ramaria pallida | T |
| 1 | Rhodocollybia spp. | * |
| 1 | Russula foetens | T |
| 1 | Russula persicina | NE |
| 1 | Russula torulosa | T |
| 1 | Suillus collinitus | E |
| 1 | Tricholoma equestre | T |
| Total: 448 |
| Species | Edibility | Specimens (n) | ITS Ref. Seq. (n) | |
|---|---|---|---|---|
| building | Amanita caesarea | E | 1 | 1 |
| Cantharellus ferruginascens | E | 2 | 2 | |
| Cortinarius semisanguifluus | M | 3 | 3 | |
| Desarmillaria tabescens | E | 1 | 1 | |
| Macrolepiota venenata | T | 2 | 2 | |
| Russula vinosobrunnea | E | 1 | 1 | |
| Tricholoma basirubens | E | 1 | 1 | |
| Tricholoma bresadolanum | T | 1 | 1 | |
| Tricholoma gausapatum | E | 2 | 2 | |
| Tricholoma quercicola | SNE | 1 | 1 | |
| TOT. | 10 | 15 | 15 | |
| validation | Agaricus bresadolanus | T | 2 | 2 |
| Agaricus menieri | T | 1 | 1 | |
| Amanita citrina | T | 1 | 1 | |
| Amanita excelsa | SNE | 2 | 2 | |
| Amanita muscaria | T | 1 | 1 | |
| Amanita ovoidea | T | 1 | 1 | |
| Amanita pantherina | T | 1 | 1 | |
| Amanita phalloides | M | 1 | 1 | |
| Amanita strobiliformis | E | 1 | 1 * | |
| Boletus edulis | E | 2 | 2 | |
| Butyriboletus pseudoregius | E | 1 | 1 | |
| Clitocybe phaeophtalma | T | 1 | 1 | |
| Clitopaxillus alexandri | SNE | 2 | 2 * | |
| Clitopilus prunulus | E | 1 | 1 | |
| Cortinarius cedretorum | NE | 1 | 1 | |
| Cortinarius orellanus | M | 1 | 1 | |
| Cyclocybe cylindracea | E | 1 | 1 | |
| Entoloma lividoalbum | SNE | 2 | 2 | |
| Entoloma sinuatum | T | 1 | 1 | |
| Gymnopus erythropus | NE | 1 | 1 | |
| Gyroporus castaneus | T | 1 | 1 | |
| Infundibulicybe geotropa | E | 1 | 1 | |
| Infundibulicybe mediterranea | E | 3 | 3 | |
| Inocybe geophylla | T | 1 | 1 | |
| Lactarius blennius | T | 1 | 1 | |
| Lactarius chrysorreus | NE | 1 | 1 | |
| Lactarius decipiens | NE | 1 | 1 | |
| Lactarius pyrogalus | NE | 1 | 1 | |
| Lactarius quietus | NE | 1 | 1 | |
| Lactarius torminosus | T | 1 | 1 | |
| Lactarius zonarius | T | 1 | 1 | |
| Lepiota brunneoincarnata | M | 1 | 1 | |
| Lepiota cristata | T | 1 | 1 | |
| Lepiota clypeolaria | T | 2 | 2 | |
| Lepiota ignivolvata | T | 1 | 1 | |
| Lepista glaucocana | E | 1 | 1 ** | |
| Lepista nuda | E | 1 | 1 | |
| Lepista sordida | NE | 1 | 1 | |
| Leucoagaricus leucothites | T | 1 | 1 | |
| Macrolepiota permixta | E | 1 | 1 ** | |
| Macrolepiota procera | E | 1 | 1 | |
| Neoboletus erythropus | E | 1 | 1 ** | |
| Omphalotus olearius | T | 1 | 1 | |
| Parasola conopilea | T | 2 | 2 | |
| Pholiota mutabilis | E | 1 | 1 * | |
| Ramaria flavescens | E | 2 | 2 | |
| Ramaria formosa | T | 1 | 1 | |
| Ramaria pallida | T | 1 | 1 * | |
| Rubroboletus satanas | T | 1 | 1 ** | |
| Russula caerulea | SNE | 2 | 2 | |
| Russula chloroides | NE | 1 | 1 | |
| Russula heterophylla | E | 1 | 1 | |
| Russula nobilis | NE | 2 | 2 | |
| Russula persicina | NE | 2 | 2 | |
| Russula queletii | T | 1 | 1 | |
| Russula sanguinea | NE | 1 | 1 | |
| Russula torulosa | T | 2 | 2 | |
| Russula vesca | E | 1 | 1 | |
| Scleroderma polyrhizum | T | 1 | 1 | |
| Suillus grevillei | E | 1 | 1 | |
| Suillus viscidus | E | 1 | 1 | |
| Tricholoma fracticum | T | 2 | 2 | |
| Tricholoma scalpturatum | E | 1 | 1 | |
| Tricholoma sciodes | T | 2 | 2 | |
| Tricholoma sejunctum | T | 1 | 1 | |
| Tricholoma sulphureum | T | 1 | 1 | |
| TOTAL | 66 | 82 | 82 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giusti, A.; Ricci, E.; Gasperetti, L.; Galgani, M.; Polidori, L.; Verdigi, F.; Narducci, R.; Armani, A. Building of an Internal Transcribed Spacer (ITS) Gene Dataset to Support the Italian Health Service in Mushroom Identification. Foods 2021, 10, 1193. https://doi.org/10.3390/foods10061193
Giusti A, Ricci E, Gasperetti L, Galgani M, Polidori L, Verdigi F, Narducci R, Armani A. Building of an Internal Transcribed Spacer (ITS) Gene Dataset to Support the Italian Health Service in Mushroom Identification. Foods. 2021; 10(6):1193. https://doi.org/10.3390/foods10061193
Chicago/Turabian StyleGiusti, Alice, Enrica Ricci, Laura Gasperetti, Marta Galgani, Luca Polidori, Francesco Verdigi, Roberto Narducci, and Andrea Armani. 2021. "Building of an Internal Transcribed Spacer (ITS) Gene Dataset to Support the Italian Health Service in Mushroom Identification" Foods 10, no. 6: 1193. https://doi.org/10.3390/foods10061193
APA StyleGiusti, A., Ricci, E., Gasperetti, L., Galgani, M., Polidori, L., Verdigi, F., Narducci, R., & Armani, A. (2021). Building of an Internal Transcribed Spacer (ITS) Gene Dataset to Support the Italian Health Service in Mushroom Identification. Foods, 10(6), 1193. https://doi.org/10.3390/foods10061193