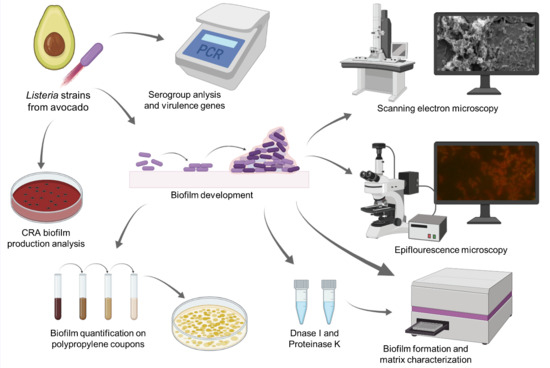
Conditions of In Vitro Biofilm Formation by Serogroups of Listeria monocytogenes Isolated from Hass Avocados Sold at Markets in Mexico
Abstract
:1. Introduction
2. Materials and Methods
2.1. L. monocytogenes Isolates
2.2. PCR-Serogroup Analysis and Virulence Genes
2.3. Biofilm Formation Assay
2.4. Conditions and Quantification of Mono-Species Biofilms
2.5. Epifluorescence Microscopy and Scanning Electron Microscopy (SEM)
2.6. Determination of Biofilm Components
2.6.1. Matrix Characterization
2.6.2. Phenotype Analysis of Biofilm Production
2.7. Statistical Analysis
3. Results
3.1. Prevalence of Virulence Genes and Distribution of L. monocytogenes Serogroups Based on PCR Analysis
3.2. Ability to Form Biofilms
3.3. Quantification and Components of the Matrix Biofilm Formation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vázquez-Boland, J.A.; Kuhn, M.; Berche, P.; Chakraborty, T.; Domínguez-Bernal, G.; Goebel, W.; González-Zorn, B.; Wehland, J.; Kreft, J. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 2001, 14, 584–640. [Google Scholar] [CrossRef] [Green Version]
- Charlier, C.; Perrodeau, É.; Leclercq, A.; Cazenave, B.; Pilmis, B.; Henry, B.; Lopes, A.; Maury, M.; Moura, A.; Goffinet, F.; et al. Clinical features and prognostic factors of listeriosis: The MONALISA national prospective cohort study. Lancet Infect. Dis. 2017, 17, 510–519. [Google Scholar] [CrossRef]
- Smith, A.; Hearn, J.; Taylor, C.; Wheelhouse, N.; Kaczmarek, M.; Moorhouse, E.; Singleton, I. Listeria monocytogenes isolates from ready to eat plant produce are diverse and have virulence potential. Int. J. Food Microbiol. 2019, 299, 23–32. [Google Scholar] [CrossRef] [Green Version]
- CDC (Center for Disease Control and Prevention). Listeria Outbreak Linked to Fully Cooked Chicken. 2021. Available online: https://www.cdc.gov/listeria/outbreaks/precooked-chicken-07-21/index.html (accessed on 31 July 2021).
- Dewey-Mattia, D.; Manikonda, K.; Hall, A.J.; Wise, M.E.; Crowe, S.J. Surveillance for Foodborne Disease Outbreaks—United States, 2009–2015. MMWR Surveill. Summ. 2018, 67, 1–11. [Google Scholar] [CrossRef]
- IFSAC (Interagency Food Safety Analytics Collaboration). Foodborne Illness Source Attribution Estimates for 2018 Salmonella, Escherichia coli, Listeria monocytogenes, and Campylobacter Using Multi-Year Outbreak Surveillance Date, United States. Atlanta, Georgia and Washington, District of Columbia: U.S. Department of Health and Human Services, CDC, FDA, USDA/FSIS. 2000. Available online: https://www.cdc.gov/foodsafety/ifsac/pdf/P19-2018-report-TriAgency-508.pdf (accessed on 2 August 2021).
- CDC (Center for Disease Control and Prevention). List of Selected Multistate Foodborne Outbreak Investigations. 2021. Available online: https://www.cdc.gov/foodsafety/outbreaks/multistate-outbreaks/outbreaks-list.html (accessed on 2 August 2021).
- FDA (Food Drugs and Administration). Microbiological Surveillance Sampling: FY18-21 Fresh Herbs (Cilantro, Basil & Parsley) and Processed Avocado and Guacamole Assignments. 2021. Available online: https://www.fda.gov/food/sampling-protect-food-supply/microbiological-surveillance-sampling-fy18-19-fresh-herbs-cilantro-basil-parsley-and-processed (accessed on 2 August 2021).
- Hoelzer, K.; Oliver, H.F.; Kohl, L.R.; Hollingsworth, J.; Wells, M.T.; Wiedmann, M. Structured expert elicitation about Listeria monocytogenes scross-contamination in the environment of retail deli operations in the United States. Risk Anal. 2012, 32, 1139–1156. [Google Scholar] [CrossRef]
- Scollon, A.M.; Wang, H.; Ryser, E.T. Transfer of Listeria monocytogenes during mechanical slicing of onions. Food Control 2016, 65, 160–167. [Google Scholar] [CrossRef] [Green Version]
- Smith, A.; Moorhouse, E.; Monaghan, J.; Taylor, C.; Singleton, I. Sources and survival of Listeria monocytogenes on fresh, leafy produce. J. Appl. Microbiol. 2018, 125, 930–942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Frutos, R.; Martínez-Chávez, L.; Cabrera-Díaz, E.; Gutiérrez-González, P.; Montañez-Soto, J.L.; Varela-Hernández, J.J.; Martínez-Gonzáles, N.E. Salmonella, Listeria monocytogenes, and indicator microorganisms on hass avocados sold at retail markets in Guadalajara, Mexico. J. Food Prot. 2020, 83, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, M.; Wang, J.; Wu, Q.; Cheng, J.; Zhang, J.; Sun, Q.; Xue, L.; Zeng, H.; Lei, T.; et al. Heterogeneity, characteristics, and public health implications of Listeria monocytogenes in ready-to-eat foods and pasteurized milk in China. Front. Microbiol. 2020, 11, 642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doumith, M.; Buchrieser, C.; Glaser, P.; Jacquet, C.; Martin, P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 2004, 42, 3819–3822. [Google Scholar] [CrossRef] [Green Version]
- Vázquez-Sánchez, D.; Galvão, J.A.; Oetterer, M. Contamination sources, serogroups, biofilm-forming ability and biocide resistance of Listeria monocytogenes persistent in tilapia-processing facilities. J. Food Sci. Technol. 2017, 54, 3867–3879. [Google Scholar] [CrossRef]
- Yin, Y.; Yao, H.; Doijad, S.; Kong, S.; Shen, Y.; Cai, X.; Tan, W.; Wang, Y.; Feng, Y.; Ling, Z.; et al. A hybrid sub-lineage of Listeria monocytogenes comprising hypervirulent isolates. Nature 2019, 10, 4283. [Google Scholar] [CrossRef] [Green Version]
- Beresford, M.R.; Andrew, P.W.; Shama, G. Listeria monocytogenes adheres to many materials found in food-processing environments. J. Appl. Microbiol. 2001, 90, 1000–1005. [Google Scholar] [CrossRef] [PubMed]
- Colagiorgi, A.; Di Ciccio, P.; Zanardi, E.; Ghidini, S.; Ianieri, A. A Look inside the Listeria monocytogenes Biofilms Extracellular Matrix. Microorganisms 2016, 4, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piercey, M.J.; Hingston, P.A.; Truelstrup, H.L. Genes involved in Listeria monocytogenes biofilm formation at a simulated food processing plant temperature of 15 °C. Int. J. Food Microbiol. 2016, 223, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, R.L.; Gorris, L.G.M.; Hayman, M.M.; Jackson, T.C.; Whiting, R.C. A review of Listeria monocytogenes: An update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control 2017, 75, 1–13. [Google Scholar] [CrossRef]
- Sadekuzzaman, M.; Yang, S.; Mizan, M.F.R.; Kim, H.S.; Ha, S.D. Effectiveness of a phage cocktail as a biocontrol agent against L. monocytogenes biofilms. Food Control 2017, 78, 256–263. [Google Scholar] [CrossRef]
- Borucki, M.K.; Peppin, J.D.; White, D.; Loge, F.; Call, D.R. Variation in Biofilm Formation among Strains of Listeria monocytogenes. Appl. Environ. Microbiol. 2003, 69, 7336–7342. [Google Scholar] [CrossRef] [Green Version]
- Flemming, H.C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Avila-Novoa, M.G.; Guerrero-Medina, P.J.; Navarrete-Sahagún, V.; Gómez-Olmos, I.; Velázquez-Suárez, N.Y.; De la Cruz-Color, L.; Gutiérrez-Lomelí, M. Biofilm formation by multidrug-resistant serotypes of Salmonella isolated from fresh products: Effects of nutritional and environmental conditions. Appl. Sci. 2021, 11, 3581. [Google Scholar] [CrossRef]
- Montero, D.; Bodero, M.; Riveros, G.; Lapierre, L.; Gaggero, A.; Vidal, R.M.; Vidal, M. Molecular epidemiology and genetic diversity of Listeria monocytogenes isolates from a wide variety of ready-to-eat foods and their relationship to clinical strains from listeriosis outbreaks in Chile. Front. Microbiol. 2015, 6, 384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stepanović, S.; Ćirković, I.; Ranin, L.; Švabić-Vlahović, M. Biofilm formation by Salmonella spp. and Listeria monocytogenes on plastic surface. Lett. Appl. Microbiol. 2004, 38, 428–432. [Google Scholar] [CrossRef]
- In Lee, S.H.; Barancelli, G.V.; de Camargo, T.M.; Corassin, C.H.; Rosim, R.E.; Gomes, A.C.; Pereira, C.L.; Fernandes, C.A.O. Biofilm-producing ability of Listeria monocytogenes isolates from Brazilian cheese processing plants. Food Res. Int. 2017, 91, 88–91. [Google Scholar] [CrossRef] [PubMed]
- Fratesi, S.E.; Lynch, F.L.; Kirkland, B.L.; Brown, L.R. Effects of SEM Preparation Techniques on the Appearance of Bacteria and Biofilms in the Carter Sandstone. J. Sediment. Res. 2004, 74, 858–867. [Google Scholar] [CrossRef]
- Fredheim, E.G.A.; Klingenberg, C.; Rohde, H.; Frankenberger, S.; Gaustad, P.; Flægstad, T.; Sollid, J.E. Biofilm formation by Staphylococcus haemolyticus. J. Clin. Microbiol. 2009, 47, 1172–1180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avila-Novoa, M.G.; González-Gómez, J.P.; Guerrero-Medina, P.J.; Cardona-López, M.A.; Ibarra-Velazquez, L.M.; Velazquez-Suarez, N.Y.; Morales-Del Río, J.A.; Gutiérrez-Lomelí, M. Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) strains isolated from dairy products: Relationship of ica-dependent/independent and components of biofilms produced in vitro. Int. Dairy J. 2021, 119, 105066. [Google Scholar] [CrossRef]
- Arciola, C.R.; Baldassarri, L.; Montanaro, L. Presence of icaA and icaD genes and slime production in a collection of staphylococcal strains from catheter-associated infections. J. Clin. Microbiol. 2001, 39, 2151–2156. [Google Scholar] [CrossRef] [Green Version]
- Han, Q.; Song, X.; Zhang, Z.; Fu, J.; Wang, X.; Malakar, P.K.; Lui, H.; Pan, Y.; Zhao, Y. Removal of foodborne pathogen biofilms by acidic electrolyzed water. Front. Microbiol. 2017, 8, 988. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Zhao, F.; Wang, J.; Zhong, N. Biofilm formation and control strategies of foodborne pathogens: Food safety perspectives. RSC Adv. 2017, 7, 36670–36683. [Google Scholar] [CrossRef] [Green Version]
- Korsak, D.; Borek, A.; Daniluk, S.; Grabowska, A.; Pappelbaum, K. Antimicrobial susceptibilities of Listeria monocytogenes strains isolated from food and food processing environment in Poland. Int. J. Food Microbiol. 2012, 158, 203–208. [Google Scholar] [CrossRef]
- Laksanalamai, P.; Huang, B.; Sabo, J.; Burall, L.S.; Zhao, S.; Bates, J.; Datta, A.R. Genomic characterization of novel Listeria monocytogenes serotype 4b variant strains. PLoS ONE 2014, 9, 89024. [Google Scholar] [CrossRef] [Green Version]
- Lomonaco, S.; Verghese, B.; Gerner-Smidt, P.; Tarr, C.; Gladney, L.; Joseph, L.; Katz, L.; Turnsek, M.; Frace, M.; Chen, Y.; et al. Novel Epidemic Clones of Listeria United States. Emerg. Infect. Dis. 2013, 19, 147–150. [Google Scholar] [CrossRef]
- Roche, S.M.; Grépinet, O.; Kerouanton, A.; Ragon, M.; Leclercq, A.; Témoin, S.; Schaeffer, B.; Skorski, G.; Mereghetti, L.; Le Monnier, A.; et al. Polyphasic characterization and genetic relatedness of low-virulence and virulent Listeria monocytogenes isolates. BMC Microbiol. 2012, 12, 304. [Google Scholar] [CrossRef] [Green Version]
- Vilchis-Rangel, R.E.; Espinoza-Mellado, M.R.; Salinas-Jaramillo, I.J.; Martinez-Peña, M.D.; Rodas-Suárez, O.R. Association of Listeria monocytogenes LIPI-1 and LIPI-3 marker llsX with invasiveness. Curr. Microbiol. 2019, 76, 637–643. [Google Scholar] [CrossRef]
- Su, X.; Zhang, J.; Shi, W.; Yang, X.; Li, Y.; Pan, H.; Kuang, D.; Xu, X.; Shi, X.; Meng, J. Molecular characterization and antimicrobial susceptibility of Listeria monocytogenes isolated from foods and humans. Food Control 2016, 70, 96–102. [Google Scholar] [CrossRef]
- Osman, K.M.; Kappell, A.D.; Fox, E.M.; Orabi, A.; Samir, A. Prevalence, pathogenicity, virulence, antibiotic resistance, and phylogenetic analysis of biofilm producing Listeria monocytogenes isolated from different ecological niches in Egypt: Food, humans, animals, and environment. Pathogens 2019, 9, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López, V.; Navas, J.; Martínez-Suárez, J.V. Low potential virulence associated with mutations in the inla and prfa genes in Listeria monocytogenes isolated from raw retail poultry meat. J. Food Prot. 2013, 76, 129–132. [Google Scholar] [CrossRef]
- Kumar, S.; Parvathi, A.; George, J.; Krohne, G.; Karunasagar, I.; Karunasagar, I. A study on the effects of some laboratory-derived genetic mutations on biofilm formation by Listeria monocytogenes. World J. Microbiol. Biotechnol. 2009, 25, 527–531. [Google Scholar] [CrossRef]
- Scortti, M.; Monzó, H.J.; Lacharme-Lora, L.; Lewis, D.A.; Vázquez-Boland, J.A. The PrfA virulence regulon. Microbes Infect. 2007, 9, 1196–1207. [Google Scholar] [CrossRef] [PubMed]
- Osman, K.M.; Samir, A.; Abo-Shama, U.H.; Mohamed, E.H.; Orabi, A.; Zolnikov, T. Determination of virulence and antibiotic resistance pattern of biofilm producing Listeria species isolated from retail raw milk. BMC Microbiol. 2016, 16, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kadam, S.R.; den Besten, H.M.W.; van der Veen, S.; Zwietering, M.H.; Moezelaar, R.; Abee, T. Diversity assessment of Listeria monocytogenes biofilm formation: Impact of growth condition, serotype and strain origin. Int. J. Food Microbiol. 2013, 165, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.; Wolfaardt, G.M.; Arts, M.T. Characterization of pseudomonas aeruginosa fatty acid profiles in biofilms and batch planktonic cultures. Can. J. Microbiol. 2010, 56, 1028–1039. [Google Scholar] [CrossRef] [Green Version]
- Perez, L.J.; Ng, W.L.; Marano, P.; Brook, K.; Bassler, B.L.; Semmelhack, M.F. Role of the CAI-1 fatty acid tail in the Vibrio cholerae quorum sensing response. J. Med. Chem. 2012, 55, 9669–9681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keeney, K.; Trmcic, A.; Zhu, Z.; Delaquis, P.; Wang, S. Stress survival islet 1 contributes to serotype-specific differences in biofilm formation in Listeria monocytogenes. Lett. Appl. Microbiol. 2018, 67, 530–536. [Google Scholar] [CrossRef]
- De Oliveira, M.M.; Brugnera, D.F.; Alves, E.; Piccoli, R.H. Biofilm formation by Listeria monocytogenes on stainless steel surface and biotransfer potential. Braz. J. Microbiol. 2010, 41, 97–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Djordjevic, D.; Wiedmann, M.; McLandsborough, L.A. Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl. Environ. Microbiol. 2002, 68, 2950–2958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, H.; Miya, S.; Igarashi, K.; Suda, T.; Kuramoto, S.; Kimura, B. Biofilm formation ability of Listeria monocytogenes isolates from raw ready-to-eat seafood. J. Food Prot. 2009, 72, 1476–1480. [Google Scholar] [CrossRef]
- Pantanella, F.; Valenti, P.; Natalizi, T.; Passeri, D.; Berlutti, F. Analytical techniques to study microbial biofilm on abiotic surfaces: Pros and cons of the main techniques currently in use. Ann Ig 2013, 25, 31–42. [Google Scholar]
- Azeredo, J.; Azevedo, N.F.; Briandet, R.; Cerca, N.; Coenye, T.; Costa, A.R.; Desvaux, M.; Di Bonaventura, G.D.; Hébraud, M.; Jaglic, Z.; et al. Critical review on biofilm methods. Crit. Rev. Microbiol. 2017, 43, 313–351. [Google Scholar] [CrossRef] [Green Version]
- Di Bonaventura, G.; Piccolomini, R.; Paludi, D.; D’Orio, V.; Vergara, A.; Conter, M.; Ianieri, A. Influence of temperature on biofilm formation by Listeria monocytogenes on various food-contact surfaces: Relationship with motility and cell surface hydrophobicity. J. Appl. Microbiol. 2008, 104, 1552–1561. [Google Scholar] [CrossRef]
- Pan, Y.; Breidt, F.; Kathariou, S. Competition of Listeria monocytogenes serotype 1/2a and 4b strains in mixed-culture biofilms. Appl. Environ. Microbiol. 2009, 75, 5846–5852. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, R.E.; Ross, T.; Bowman, J.P. Variability in biofilm production by Listeria monocytogenes correlated to strain origin and growth conditions. Int. J. Food Microbiol. 2011, 150, 14–24. [Google Scholar] [CrossRef]
- Kocot, A.M.; Olszewska, M.A. Biofilm formation and microscopic analysis of biofilms formed by Listeria monocytogenes in a food processing context. LWT 2017, 84, 47–57. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Renier, S.; Hébraud, M.; Desvaux, M. Molecular biology of surface colonization by Listeria monocytogenes: An additional facet of an opportunistic Gram-positive foodborne pathogen. Environ. Microbiol. 2011, 13, 835–850. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Cody, G.D.; Harding, A.K.; Wilmes, P.; Schrenk, M.; Wheeler, K.E.; Banfield, J.F.; Thelen, M.P. Characterization of extracellular polymeric substances from acidophilic microbial biofilms. Appl. Environ. Microbiol. 2010, 76, 2916–2922. [Google Scholar] [CrossRef] [Green Version]
- Muthukrishnan, G.; Quinn, G.A.; Lamers, R.P.; Diaz, C.; Cole, L.; Chen, S.; Cole, A.M. Exoproteome of Staphylococcus aureus reveals putative determinants of nasal carriage. J. Proteome Res. 2011, 10, 2064–2078. [Google Scholar] [CrossRef] [Green Version]
- Harmsen, M.; Lappann, M.; Knøchel, S.; Molin, S. Role of extracellular DNA during biofilm formation by Listeria monocytogenes. Appl. Environ. Microbiol. 2010, 76, 2271–2279. [Google Scholar] [CrossRef] [Green Version]
- Okshevsky, M.; Meyer, R.L. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit. Rev. Microbiol. 2015, 41, 341–352. [Google Scholar] [CrossRef]
- Alhede, M.; Bjarnsholt, T.; Givskov, M.; Alhede, M. Pseudomonas aeruginosa biofilms. Mechanisms of immune evasion. Adv. Appl. Microbiol. 2014, 86, 1–40. [Google Scholar]
- Ibáñez de Aldecoa, A.L.; Zafra, O.; González-Pastor, J.E. Mechanisms and regulation of extracellular DNA release and its biological roles in microbial communities. Front. Microbiol. 2017, 8, 1390. [Google Scholar] [CrossRef] [Green Version]
- Thévenot, D.; Dernburg, A.; Vernozy-Rozand, C. An updated review of Listeria monocytogenes in the pork meat industry and its products. J. Appl. Microbiol. 2006, 101, 7–17. [Google Scholar] [CrossRef]
- Araújo, P.A.; Lemos, M.; Mergulhão, F.; Melo, L.; Simões, M. The influence of interfering substances on the antimicrobial activity of selected quaternary ammonium compounds. Int. J. Food Sci. 2013, 2013, 237581. [Google Scholar] [CrossRef] [Green Version]
- Chiang, W.C.; Nilsson, M.; Jensen, P.Ø.; Høiby, N.; Nielsen, T.E.; Givskov, M.; Tolker-Nielsen, T. Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 2013, 57, 2352–2361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaitiemwong, N.; Hazeleger, W.C.; Beumer, R.R. Inactivation of Listeria monocytogenes by disinfectants and bacteriophages in suspension and stainless steel carrier tests. J. Food Prot. 2014, 77, 2012–2020. [Google Scholar] [CrossRef] [PubMed]
- Lutskiy, M.Y.; Avneri-Katz, S.; Zhu, N.; Itsko, M.; Ronen, Z.; Arnusch, C.J.; Kasher, R. A microbiology-based assay for quantification of bacterial early stage biofilm formation on reverse-osmosis and nanofiltration membranes. Sep. Purif. Technol. 2015, 141, 214–220. [Google Scholar] [CrossRef]



| Primers | Length (bp) | Sequence 5′—3′ | Tm (°C) | |
|---|---|---|---|---|
| plcB | 723 | Forward | 5′-CAG CTC CGC ATG ATA TTG AC-3′ | 58 |
| Reverse | 5′-CTG CCA AAG TTT GCT GTG AA-3′ | |||
| Inl 1A | 629 | Forward | 5′-GGC TGG GCA TAA CCA AAT TA-3′ | 60 |
| Reverse | 5′-CTT TTG GTG CCG TAG GT-3′ | |||
| Inl 1B | 293 | Forward | 5′-CCT AAA CCT CCG ACC AAA CA-3′ | 60 |
| Reverse | 5′-CCA TTT CGC GCT TCT CTA TC-3′ | |||
| prfA | 330 | Forward | 5′-ACC AAT GGG ATC CAC AAG AA-3′ | 58 |
| Reverse | 5′-GCT TCC CGT TAA TCG AAA AAT-3′ | |||
| icA | 840 | Forward | 5′-TCC CAT TAG GTG GAA AAG CA-3′ | 57 |
| plcA | Reverse | 5′-CGG GGA AGT CCA TGA TTA GA-3′ | ||
| Hly | 1100 | Forward | 5′-GTC TAC CAA TTG CGC AAC AA-3′ | 57 |
| Reverse | 5′-TGG TGT TTC CCG GTT AAA AG-3′ | |||
| Mpl | 450 | Forward | 5′-AAA GGT GGA GAA ATT GAT TCG-3′ | 62 |
| Reverse | 5′-AGT GAT CGT ATT GTA GGC TGC TT-3′ | |||
| actA | 571 | Forward | 5′-AAA CAG AAG AGC CAA GC-3′ | 58 |
| Reverse | 5′-TTC ACT TCG GGA TTT TCG TC-3′ | |||
| PCR-Serogroups | Strain ID | Genetic Determinants of Virulence No. of Isolates (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| prfA | plcA | hly | mpl | actA | plcB | inlB | inlA | ||
| I | Lm-334 | + | − | + | + | + | − | − | + |
| III | Lm-132 | − | − | − | − | − | − | + | − |
| Lm-237 | − | + | − | + | + | − | + | − | |
| Lm-249 | + | + | + | + | + | − | + | − | |
| Lm-252 | − | + | + | + | + | − | + | + | |
| Lm-253 | + | + | + | + | + | − | + | − | |
| Lm-304 * | + | + | + | + | + | + | + | − | |
| Lm-333 * | + | + | + | + | + | + | + | − | |
| Lm-335 | + | + | − | + | + | + | − | − | |
| Lm-336 * | + | + | + | + | + | + | + | + | |
| Lm-352 * | + | + | + | + | + | + | − | + | |
| Lm-356 * | + | + | + | + | + | + | + | − | |
| Lm-357 | + | + | + | − | + | + | + | − | |
| IV | Lm-142 | − | + | − | − | − | − | − | − |
| Lm-251 | − | + | + | + | + | - | + | + | |
| Lm-303 * | + | + | + | + | + | + | + | − | |
| Lm-320 * | + | + | + | + | + | + | + | − | |
| Lm-332 | − | + | + | + | + | + | + | + | |
| Total | 18 (100%) | 12 (66.6%) | 16 (88.88%) | 14 (77.7%) | 15 (83.3%) | 16 (88.8%) | 10 (55.5%) | 14 (77.7%) | 6 (33.3%) |
| Time | Medium Culture | No. of Strains (%) Polystyrene Microtiter Plates Biofilm Formation | |||
|---|---|---|---|---|---|
| Strong Biofilm | Moderate Biofilm | Weak Biofilm | Non-Biofilm | ||
| Producers | Producers | Producers | Producers | ||
| 48 h | TSBAa | 4 (22.2) | 0 | 7 (38.8) | 7 (38.8) |
| OD570 mean = 0.346 ± 0.007 | OD570 mean = 0.091 ± 0.007 | OD570 mean = 0.075 ± 0.000 | |||
| 1/10 diluted TSBAb | 4 (22.2) | 0 | 14 (77.7) | 0 | |
| OD570 mean = 0.289 ± 0.002 | OD570 mean = 0.076 ± 0.004 | ||||
| TSBc | 0 | 1 (5.5) | 17 (94.4) | 0 | |
| OD570 mean = 0.210 ± 0.014 | OD570 mean = 0.093 ± 0.018 | ||||
| 1/10 diluted TSBd | 0 | 4 (22.2) | 14 (77.7) | 0 | |
| OD570 mean = 0.125 ± 0.006 | OD570 mean = 0.072 ± 0.002 | ||||
| 240 h | TSBAa | 6 (33.3) | 12 (66.6) | 0 | 0 |
| OD570 mean = 0.402 ± 0.078 | OD570 mean = 0.218 ± 0.082 | ||||
| 1/10 diluted TSBAb | 11 (61.1) | 7 (38.8) | 0 | 0 | |
| OD570 mean = 0.317 ± 0.013 | OD570 mean = 0.183 ± 0.018 | ||||
| TSBc | 1 (5.5) | 17 (94.4) | 0 | 0 | |
| OD570 mean = 0.302 ± 0.007 | OD570 mean = 0.226 ± 0.046 | ||||
| 1/10 diluted TSBd | 3 (16.6) | 15 (83.3) | 0 | 0 | |
| OD570 mean = 0.275 ± 0.004 | OD570 mean = 0.183 ± 0.011 | ||||
| Mono-Species Biofilms | Culture Media | Log10 CFU/cm2 ± SD A |
|---|---|---|
| Polypropylene Type B | ||
| Lm-303 | TSB | 7.54 ± 0.08 g |
| 1/10 diluted TSB | 7.52 ± 0.06 g | |
| TSBA | 8.36 ± 0.01 b | |
| 1/10 diluted TSBA | 7.54 ± 0.05 fg | |
| Lm-320 | TSB | 7.60 ± 0.08 efg |
| 1/10 diluted TSB | 7.59 ± 0.14 efg | |
| TSBA | 8.82 ± 0.03 a | |
| 1/10 diluted TSBA | 7.49 ± 0.07 g | |
| Lm-352 | TSB | 7.61 ± 0.12 defg |
| 1/10 diluted TSB | 7.56 ± 0.09 efg | |
| TSBA | 7.78 ± 0.03 cdefg | |
| 1/10 diluted TSBA | 7.66 ± 0.06 defg | |
| Lm-356 | TSB | 7.67 ± 0.10 defg |
| 1/10 diluted TSB | 7.61 ± 0.12 defg | |
| TSBA | 7.84 ± 0.30 cdef | |
| 1/10 diluted TSBA | 7.61 ± 0.11 efg | |
| Listeria monocytogenes ATCC 19111 | TSB | 7.66 ± 0.30 defg |
| 1/10 diluted TSB | 7.92 ± 0.59 cd | |
| TSBA | 8.01 ± 0.09 c | |
| 1/10 diluted TSBA | 7.85 ± 0.18 cde |
| Serogroups | Mono-Species Biofilms | Biomass Reduction (%) | |
|---|---|---|---|
| Proteinase K | DNase I | ||
| I | Lm-334 | 36.80 ± 0.10 e | 47.69 ± 0.04 a |
| III | Lm-132 | 38.98 ± 0.03 d | 7.86 ± 0.06 p |
| Lm-237 | 32.67 ± 0.07 h | 28.96 ± 0.01 e | |
| Lm-249 | 19.80 ± 0.07 m | 6.67 ± 0.00 r | |
| Lm-252 | 18.38 ± 0.04 o | 11.91 ± 0.00 m | |
| Lm-253 | 12.48 ± 0.02 q | 13.35 ± 0.17 k | |
| Lm-304 | 16.90 ± 0.11 p | 14.59 ± 0.02 j | |
| Lm-333 | 34.79 ± 0.03 g | 9.75 ± 0.04 n | |
| Lm-335 | 25.33 ± 0.01 j | 44.54 ± 0.21 c | |
| Lm-336 | 26.18 ± 0.02 i | 44.87 ± 0.06 b | |
| Lm-352 | 40.82 ± 0.07 b | 18.85 ± 0.03 g | |
| Lm-356 | 35.36 ± 0.01 f | 9.46 ± 0.00 o | |
| Lm-357 | 46.82 ± 0.06 a | 16.95 ± 0.03 h | |
| IV | Lm-142 | 39.55 ± 0.06 c | 33.08 ± 0.00 d |
| Lm-251 | 19.23 ± 0.07 n | 7.56 ± 0.03 q | |
| Lm-303 | 20.92 ± 0.03 l | 23.72 ± 0.00 f | |
| Lm-320 | 23.60 ± 0.11 k | 9.44 ± 0.0287 o | |
| Lm-332 | 16.92 ± 0.01 p | 12.83 ± 0.06 l | |
| Listeria monocytogenes ATCC 19111 | 46.80 ± 0.01 a | 15.87 ± 0.01 i | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avila-Novoa, M.G.; Navarrete-Sahagún, V.; González-Gómez, J.P.; Novoa-Valdovinos, C.; Guerrero-Medina, P.J.; García-Frutos, R.; Martínez-Chávez, L.; Martínez-Gonzáles, N.E.; Gutiérrez-Lomelí, M. Conditions of In Vitro Biofilm Formation by Serogroups of Listeria monocytogenes Isolated from Hass Avocados Sold at Markets in Mexico. Foods 2021, 10, 2097. https://doi.org/10.3390/foods10092097
Avila-Novoa MG, Navarrete-Sahagún V, González-Gómez JP, Novoa-Valdovinos C, Guerrero-Medina PJ, García-Frutos R, Martínez-Chávez L, Martínez-Gonzáles NE, Gutiérrez-Lomelí M. Conditions of In Vitro Biofilm Formation by Serogroups of Listeria monocytogenes Isolated from Hass Avocados Sold at Markets in Mexico. Foods. 2021; 10(9):2097. https://doi.org/10.3390/foods10092097
Chicago/Turabian StyleAvila-Novoa, María Guadalupe, Velia Navarrete-Sahagún, Jean Pierre González-Gómez, Carolina Novoa-Valdovinos, Pedro Javier Guerrero-Medina, Ramón García-Frutos, Liliana Martínez-Chávez, Nanci Edid Martínez-Gonzáles, and Melesio Gutiérrez-Lomelí. 2021. "Conditions of In Vitro Biofilm Formation by Serogroups of Listeria monocytogenes Isolated from Hass Avocados Sold at Markets in Mexico" Foods 10, no. 9: 2097. https://doi.org/10.3390/foods10092097
APA StyleAvila-Novoa, M. G., Navarrete-Sahagún, V., González-Gómez, J. P., Novoa-Valdovinos, C., Guerrero-Medina, P. J., García-Frutos, R., Martínez-Chávez, L., Martínez-Gonzáles, N. E., & Gutiérrez-Lomelí, M. (2021). Conditions of In Vitro Biofilm Formation by Serogroups of Listeria monocytogenes Isolated from Hass Avocados Sold at Markets in Mexico. Foods, 10(9), 2097. https://doi.org/10.3390/foods10092097