The Influence of DNA Extraction Methods on Species Identification Results of Seafood Products
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Sample Processing
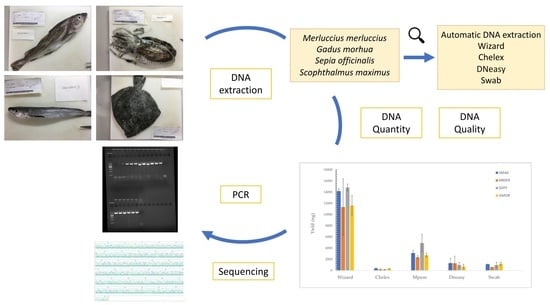
2.3. DNA Extraction
2.3.1. Wizard DNA Clean-Up System Kit (Promega, Madison, WI, USA)
2.3.2. MPure-12TM Automated Nucleic Acid Purification System (MP Biomedicals, Santa Ana, CA, USA)
2.3.3. Chelex 100 resin (Biorad, Hercules, CA, USA)
2.3.4. DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany)
2.3.5. Swab Method
2.4. DNA Quantity and Quality Determination
2.4.1. Yield and Efficiency
2.4.2. Purity
2.4.3. Integrity of Extracted DNA
2.5. Handling Time and Total Extraction Time
2.6. PCR Amplification and Sequencing
2.6.1. PCR
2.6.2. Sanger Sequencing
2.6.3. Sequence Quality Determination
2.6.4. Species Authentication
2.7. Complementary Parameters
2.7.1. Safety
2.7.2. Affordability
2.7.3. Simplicity
2.7.4. Automation
2.8. Statistical Analyses
3. Results
3.1. DNA Quantity and Quality
3.1.1. Yield and Efficiency
3.1.2. Purity
3.1.3. Integrity of Extracted DNA
3.2. Handling Time and Total Extraction Time
3.3. PCR and Sequencing
3.4. Complementary Parameters
4. Discussion
4.1. DNA Quantity and Quality
4.2. Handling Time and Total Extraction Time
4.3. PCR and Sequencing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dhaliwal, A. DNA extraction and purification. Mater Methods 2013, 3, 191. [Google Scholar] [CrossRef]
- Di Pinto, A.; Forte, V.; Guastadisegni, M.C.; Martino, C.; Schena, F.P.; Tantillo, G. A comparison of DNA extraction methods for food analysis. Food Control. 2007, 18, 76–80. [Google Scholar] [CrossRef]
- Armani, A.; Tinacci, L.; Xiong, X.; Titarenko, E.; Guidi, A.; Castigliego, L. Development of a Simple and Cost-Effective Bead-Milling Method for DNA Extraction from Fish Muscles. Food Anal. Methods 2014, 7, 946–955. [Google Scholar]
- Dairawan, M.; Shett, P.J. The evolution of DNA extraction methods. Am. J. Biomed. Sci. Res. 2020, 8, 39–46. [Google Scholar]
- Lopera-Barrero, N.M.; Povh, J.A.; Ribeiro, R.P.; Gomes, P.C.; Jacometo, C.B.; Silva Lopes, T.D. Comparación de protocolos de extracción de ADN con muestras de aleta y larva de peces: Extracción modificada con cloruro de sodio. Cienc. E Investig. Agrar. 2008, 35, 77–86. [Google Scholar] [CrossRef] [Green Version]
- Pardo, M.A.; Pérez-Villareal, B. Identification of commercial canned tuna species by restriction site analysis of mitochondrial DNA products obtained by nested primer PCR. Food Chem. 2004, 86, 143–150. [Google Scholar] [CrossRef]
- Chapela, M.J.; Sotelo, C.G.; Pérez-Martín, R.I.; Pardo, M.Á.; Pérez-Villareal, B.; Gilardi, P.; Riese, J. Comparison of DNA extraction methods from muscle of canned tuna for species identification. Food Control. 2007, 18, 1211–1215. [Google Scholar] [CrossRef]
- Tan, S.C.; Yiap, B.C. DNA, RNA, and protein extraction: The past and the present. J. Biomed. Biotechnol. 2009, 2009, 10. [Google Scholar] [CrossRef] [Green Version]
- Sepp, R.; Szabo, I.; Uda, H.; Sakamoto, H. Rapid techniques for DNA extraction from routinely processed archival tissue for use in PCR. J. Clin. Pathol. 1994, 47, 318–323. [Google Scholar] [CrossRef] [Green Version]
- Besbes, N.; Fattouch, S.; Sadok, S. Comparison of methods in the recovery and amplificability of DNA from fresh and processed sardine and anchovy muscle tissues. Food Chem. 2011, 129, 665–671. [Google Scholar] [CrossRef]
- Velasco, A.; Ramilo-Fernández, G.; Sotelo, C.G. A Real-Time PCR Method for the Authentication of Common Cuttlefish (Sepia officinalis) in Food Products. Foods 2020, 9, 286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cawthorn, D.M.; Steinman, H.A.; Witthuhn, R.C. Comparative study of different methods for the extraction of DNA from fish species commercially available in South Africa. Food Control. 2011, 22, 231–244. [Google Scholar] [CrossRef]
- Hellberg, R.S.; Kawalek, M.D.; Van, K.T.; Shen, Y.; Williams-Hill, D.M. Comparison of DNA extraction and PCR setup methods for use in high-throughput DNA barcoding of fish species. Food Anal. Methods 2014, 7, 1950–1959. [Google Scholar] [CrossRef]
- FAO Reports. The state of World Fisheries and Aquaculture 2020. Sustainability in Action; FAO: Rome, Italy, 2020. [Google Scholar]
- FAO FishFinder-The Species Identification and Data Programme. In FAO Fisheries and Aquaculture Department. Available online: http://www.fao.org/fishery/fishfinder/en (accessed on 5 April 2022).
- Programa de Control de Calidad de Ácidos Nucleicos. Banco Nacional de ADN Carlos III (Universidad de Salamanca). Available online: www.bancoadn.org (accessed on 18 March 2022).
- Sotelo, C.G.; Velasco, A.; Perez-Martin, R.I.; Kappel, K.; Schröder, U.; Verrez-Bagnis, V.; Jérôme, M.; Mendes, R.; Silva, H.; Mariani, S.; et al. Tuna labels matter in Europe: Mislabelling rates in different tuna products. PLoS ONE 2018, 13, e0196641. [Google Scholar] [CrossRef] [PubMed]
- Mariani, S.; Griffiths, A.M.; Velasco, A.; Kappel, K.; Jérôme, M.; Perez-Martin, R.I.; Schröder, U.; Verrez-Bagnis, V.; Silva, H.; Vandamme, S.G.; et al. Low mislabeling rates indicate marked improvements in European seafood market operations. Front. Ecol. Environ. 2015, 13, 536–540. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Burgener, M. Molecular Species Differentiation of Fish and Mammals. Ph.D. Thesis, University of Bern, Bern, Switzerland, 1997. [Google Scholar]
- Kumar, S.; Dudley, J.; Nei, M.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and proteinsequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef] [Green Version]
- Yalçınkaya, B.; Yumbul, E.; Mozioğlu, E.; Akgoz, M. Comparison of DNA extraction methods for meat analysis. Food Chem. 2017, 221, 1253–1257. [Google Scholar] [CrossRef]
- Besbes, N.; Sáiz-Abajo, M.J.; Sadok, S. Comparative study of DNA extraction to initiate harmonized protocol for a simple method of species identification: Fresh and canned Tuna case study. CyTA-J. Food 2022, 20, 39–49. [Google Scholar] [CrossRef]
- Singer-Sam, J.; Tanguay, R.L.; Riggs, A.D. Use of Chelex to improve the PCR signal from a small number of cells. Amplifications 1989, 3, 11. [Google Scholar]
- Meissner, H.; Fevolden, S.E.; Amundsen, P.A.; Præbel, K. Evaluation of three methods for high throughput extraction of DNA from challenging fish tissues. Conserv. Genet. Resour. 2013, 5, 733–735. [Google Scholar] [CrossRef]







| Method | Digestion Time | Handling Time | Total Extraction Time |
|---|---|---|---|
| Wizard | 2 h | 3 h | 5 h * |
| Chelex | - | 1 h 30 min | 1 h 30 min |
| DNeasy | 2 h | 2 h | 4 h |
| MPure | 2 h | 1 h | 3 h * |
| Swab | - | 1 h | 1 h |
| Sample | Wizard | Chelex | DNeasy | MPure | Swab |
|---|---|---|---|---|---|
| SMAX | + + + | − − − | + + + | + + + | − − − |
| MMER | + − + | − + − | + + + | + − + | − − − |
| GMOR | + + + | − − − | + + + | + + + | + − − |
| SOFF | + + + | + + − | + + + | + + + | − − − |
| Method | Total DNA Loaded (ng) per PCR Reaction | Sequence Quality % Burgener Primers | Sequence Quality % Folmer Primers |
|---|---|---|---|
| Wizard | 100 | 99 ± 0.5 | 93.2 ± 1.1 |
| Chelex | 11.5 ± 5.7 | 89.2 | - |
| DNeasy | 26.1 ± 19.1 | 97.3 ± 4.6 | 84 ± 15 |
| MPure | 100 | 99.2 ± 0.2 | 94.3 ± 1.3 |
| Swab | 9 ± 3.05 | - | - |
| Sample | GenBank Accession Number | Species | Query Cover (%) | % of Identity | Date of Access |
|---|---|---|---|---|---|
| SOFF1 | ON564881 | Sepia officinalis | 100 | 100 | 11/05/2022 |
| SOFF2 | ON564882 | Sepia officinalis | 100 | 100 | 11/05/2022 |
| SOFF3 | ON564883 | Sepia officinalis | 100 | 100 | 11/05/2022 |
| GMOR1 | ON505202 | Gadus morhua | 100 | 100 | 11/05/2022 |
| GMOR2 | ON505203 | Gadus morhua | 100 | 100 | 11/05/2022 |
| GMOR3 | ON505204 | Gadus morhua | 100 | 100 | 11/05/2022 |
| MMER1 | ON505205 | Merluccius merluccius | 100 | 100 | 11/05/2022 |
| MMER2 | ON505206 | Merluccius merluccius | 100 | 99.76 | 11/05/2022 |
| MMER3 | ON505207 | Merluccius merluccius | 100 | 100 | 11/05/2022 |
| SMAX1 | ON505208 | Scophthalmus maximus | 100 | 100 | 11/05/2022 |
| SMAX2 | ON505209 | Scophthalmus maximus | 100 | 100 | 11/05/2022 |
| SMAX3 | ON505210 | Scophthalmus maximus | 100 | 100 | 11/05/2022 |
| Wizard | Chelex | Dneasy | Mpure-12 | Swab | |
|---|---|---|---|---|---|
| Hazardous components and substance classification | Proteinase k solution (GHS08) Guanidinium thiocyanate (GHS05, GHS07) Isopropanol | - | Proteinase k solution (GHS08) Guanidinium chloride (H302 + H332, H315, H319), maleic acid (H302, H312, H315, H319, H317, H335) | Proteinase k solution (GHS08) Guanidinium chloride (H302 + H332, H315, H319) | - |
| Wizard | Chelex | DNeasy | MPure-12 | Swab | |
|---|---|---|---|---|---|
| Yield (Total DNA (ng)) | 11,404.72 ± 4307.16 | 194.66 ± 111.16 | 787.64 ± 721.96 | 2653.06 ± 1372.63 | 892.92 ± 336.08 |
| Efficiency (ng DNA/mg wet tissue) | 38.108 ± 14.943 | 68.393 ± 43.454 | 36.523 ± 33.292 | 66.843 ± 36.349 | 17.505 ± 7.098 |
| Purity (260/280) | 1.967 ± 0.054 | 1.843 ± 0.526 | 2.177 ± 0.263 | 2.031 ± 0.184 | 1.547 ± 0.159 |
| Rapidity (Extraction time) | 5 h * | 1 h 30′ | 4 h | 3 h * | 1 h |
| PCR amplification success % | 94.44 | 33.33 | 100 | 99.44 | 5.55 |
| Safety of components (see Table 4) | + + | + + ++ | + | + + + | + + + + |
| Affordability (reagent cost per prep) | EUR 2.22 | EUR 0.001918 | EUR 4.26 | EUR 5.65 | EUR 0.051 |
| Affordability (specific equipment value) | Vacuum manifold | Not required | Not required | MPure-12TM automated nucleic acid purification system | Not required |
| Technical simplicity | + | + + + | + + | + + | + + + + |
| Automation | - | - | - | + + + + | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Riveiro, R.; Velasco, A.; Sotelo, C.G. The Influence of DNA Extraction Methods on Species Identification Results of Seafood Products. Foods 2022, 11, 1739. https://doi.org/10.3390/foods11121739
Rodríguez-Riveiro R, Velasco A, Sotelo CG. The Influence of DNA Extraction Methods on Species Identification Results of Seafood Products. Foods. 2022; 11(12):1739. https://doi.org/10.3390/foods11121739
Chicago/Turabian StyleRodríguez-Riveiro, Rosalía, Amaya Velasco, and Carmen G. Sotelo. 2022. "The Influence of DNA Extraction Methods on Species Identification Results of Seafood Products" Foods 11, no. 12: 1739. https://doi.org/10.3390/foods11121739
APA StyleRodríguez-Riveiro, R., Velasco, A., & Sotelo, C. G. (2022). The Influence of DNA Extraction Methods on Species Identification Results of Seafood Products. Foods, 11(12), 1739. https://doi.org/10.3390/foods11121739