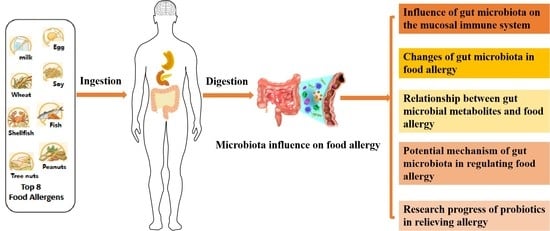
Research Progress on the Correlation between the Intestinal Microbiota and Food Allergy
Abstract
:1. Introduction
2. Food Allergy
3. Effect of Intestinal Microbiota on Mucosal Immune System
4. Influence of Dietary Intakes on Intestinal Microbiota Composition
5. Relationship between the Intestinal Microbiota and Food Allergy
5.1. Changes in the Intestinal Microbiota in Patients with Food Allergy
5.2. Relationship between Intestinal Microbial Metabolites and Food Allergy
5.3. Potential Mechanisms of Intestinal Microbiota in Regulating Food Allergy
5.3.1. Regulation of Allergen Uptake
5.3.2. Regulation of Signaling Molecules Involved in Immune Responses
6. Research Progress on the Role of Probiotics in Relieving Allergy
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Martinis, M.; Sirufo, M.M.; Suppa, M.; Ginaldi, L. New Perspectives in Food Allergy. Int. J. Mol. Sci. 2022, 21, 1474. [Google Scholar] [CrossRef]
- Peters, R.L.; Mavoa, S.; Koplin, J.J. An Overview of Environmental Risk Factors for Food Allergy. Int. J. Environ. Res. Public Health 2022, 19, 722. [Google Scholar] [CrossRef]
- Khan, M.; Sori, N. Diet-Gut Microbiota-Brain Axis and IgE-Mediated Food Allergy. Microb. Gut Brain Axis 2022, 2022, 153–168. [Google Scholar] [CrossRef]
- Peroni, D.G.; Nuzzi, G.; Trambusti, I.; Di Cicco, M.E.; Comberiati, P. Microbiome Composition and Its Impact on the Development of Allergic Diseases. Front. Immunol. 2020, 11, 700. [Google Scholar] [CrossRef]
- Joseph, C.L.; Sitarik, A.R.; Kim, H.; Huffnagle, G.; Fujimura, K.; Yong, G.J.M.; Levin, A.M.; Zoratti, E.; Lynch, S.; Ownby, D.R.; et al. Infant gut bacterial community composition and food-related manifestation of atopy in early childhood. Pediatr. Allergy Immunol. 2022, 33, e13704. [Google Scholar] [CrossRef]
- Kreft, L.; Hoffmann, C.; Ohnmacht, C. Therapeutic Potential of the Intestinal Microbiota for Immunomodulation of Food Allergies. Front. Immunol. 2020, 11, 1853. [Google Scholar] [CrossRef]
- Feehley, T.; Plunkett, C.H.; Bao, R.; Hong, S.M.C.; Culleen, E.; Belda-Ferre, P.; Campbell, E.; Aitoro, R.; Nocerino, R.; Paparo, L.; et al. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat. Med. 2019, 25, 448–453. [Google Scholar] [CrossRef]
- Pratap, K.; Taki, A.C.; Johnston, E.B.; Lopata, A.L.; Kamath, S.D. A Comprehensive Review on Natural Bioactive Compounds and Probiotics as Potential Therapeutics in Food Allergy Treatment. Front. Immunol. 2020, 11, 996. [Google Scholar] [CrossRef]
- De Martinis, M.; Sirufo, M.M.; Viscido, A.; Ginaldi, L. Food Allergy Insights: A Changing Landscape. Arch. Immunol. Ther. Exp. 2020, 68, 1–15. [Google Scholar] [CrossRef]
- Tordesillas, L.; Berin, M.C. Mechanisms of Oral Tolerance. Clin. Rev. Allergy Immunol. 2018, 55, 107–117. [Google Scholar] [CrossRef]
- Sirufo, M.M.; De Pietro, F.; Catalogna, A.; Ginaldi, L.; De Martinis, M. The Microbiota-Bone-Allergy Interplay. Int. J. Environ. Res. Public Health 2022, 19, 282. [Google Scholar] [CrossRef]
- Stephen-Victor, E.; Crestani, E.; Chatila, T.A. Dietary and Microbial Determinants in Food Allergy. Immunity 2020, 53, 277–289. [Google Scholar] [CrossRef]
- Al-Sahlany, S.T.G.; Khassaf, W.H.; Niamah, A.K.; Al-Manhel, A.J.A. Date juice addition to bio-yogurt: The effects on physicochemical and microbiological properties during storage, as well as blood parameters in vivo. J. Saudi Soc. Agric. Sci. 2022, in press. [Google Scholar] [CrossRef]
- Martens, E.C.; Neumann, M.; Desai, M.S. Interactions of commensal and pathogenic microorganisms with the intestinal mucosal barrier. Nat. Rev. Genet. 2018, 16, 457–470. [Google Scholar] [CrossRef]
- Jakobsson, H.E.; Rodríguez-Piñeiro, A.M.; Schütte, A.; Ermund, A.; Boysen, P.; Bemark, M.; Sommer, F.; Bäckhed, F.; Hansson, G.C.; Johansson, M.E.V. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep. 2015, 16, 164–177. [Google Scholar] [CrossRef]
- Kwon, O.; Han, T.-S.; Son, M.-Y. Intestinal Morphogenesis in Development, Regeneration, and Disease: The Potential Utility of Intestinal Organoids for Studying Compartmentalization of the Crypt-Villus Structure. Front. Cell Dev. Biol. 2020, 8, 593969. [Google Scholar] [CrossRef]
- Rotkiewicz, T.; Rotkiewicz, Z.; Depta, A.; Kander, M. Effect of Lactobacillus acidophilus and Bifidobacterium sp. on the course of Cryptosporidium parvum invasion in new-born piglets. Bull.-Vet. Inst. Pulawy 2001, 45, 187–196. [Google Scholar]
- Nielsen, D.S.G.; Jensen, B.B.; Theil, P.K.; Nielsen, T.S.; Knudsen, K.E.B.; Purup, S. Effect of butyrate and fermentation products on epithelial integrity in a mucus-secreting human colon cell line. J. Funct. Foods 2018, 40, 9–17. [Google Scholar] [CrossRef]
- Knudsen, K.E.B.; Lærke, H.N.; Hedemann, M.S.; Nielsen, T.S.; Ingerslev, A.K.; Nielsen, D.S.G.; Theil, P.K.; Purup, S.; Hald, S.; Schioldan, A.G.; et al. Impact of Diet-Modulated Butyrate Production on Intestinal Barrier Function and Inflammation. Nutrients 2018, 10, 1499. [Google Scholar] [CrossRef]
- Diao, H.; Jiao, A.R.; Yu, B.; Mao, X.B.; Chen, D.W. Gastric infusion of short-chain fatty acids can improve intestinal barrier function in weaned piglets. Genes Nutr. 2019, 14, 1–16. [Google Scholar] [CrossRef]
- Sonner, J.K.; Keil, M.; Falk-Paulsen, M.; Mishra, N.; Rehman, A.; Kramer, M.; Deumelandt, K.; Röwe, J.; Sanghvi, K.; Wolf, L.; et al. Dietary tryptophan links encephalogenicity of autoreactive T cells with gut microbial ecology. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, B.; Liu, X.; Fang, Y.; Zhou, P.; Zhang, Y.; Wang, Y. Flagellin as a vaccine adjuvant. Expert Rev. Vaccines 2018, 17, 335–349. [Google Scholar] [CrossRef]
- Coelho, O.G.L.; Cândido, F.G.; Alfenas, R.D.C.G. Dietary fat and gut microbiota: Mechanisms involved in obesity control. Crit. Rev. Food Sci. Nutr. 2019, 59, 3045–3053. [Google Scholar] [CrossRef]
- Agans, R.; Gordon, A.; Kramer, D.L.; Perez-Burillo, S.; Rufián-Henares, J.A.; Paliy, O. Dietary Fatty Acids Sustain the Growth of the Human Gut Microbiota. Appl. Environ. Microbiol. 2018, 84, e01525-18. [Google Scholar] [CrossRef]
- Bailén, M.; Bressa, C.; Martínez-López, S.; González-Soltero, R.; Lominchar, M.G.M.; Juan, C.S.; Larrosa, M. Microbiota Features Associated With a High-Fat/Low-Fiber Diet in Healthy Adults. Front. Nutr. 2020, 7, 583608. [Google Scholar] [CrossRef]
- Singh, R.K.; Chang, H.-W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017, 15, 73. [Google Scholar] [CrossRef] [PubMed]
- Cheng, R.; Yao, J.; Wan, Q.; Guo, J.; Pu, F.; Shi, L.; Hu, W.; Yang, Y.; Li, L.; Li, M.; et al. Oral administration of Bifidobacterium bifidum TMC3115 to neonatal mice may alleviate IgE-mediated allergic risk in adulthood. Benef. Microbes 2018, 9, 815–828. [Google Scholar] [CrossRef]
- Kostovcikova, K.; Coufal, S.; Galanova, N.; Fajstova, A.; Hudcovic, T.; Kostovcik, M.; Prochazkova, P.; Jiraskova Zakostelska, Z.; Cermakova, M.; Sediva, B.; et al. Diet Rich in Animal Protein Promotes Pro-inflammatory Macrophage Response and Exacerbates Colitis in Mice. Front. Immunol. 2019, 10, 919. [Google Scholar] [CrossRef]
- Hashemi, Z.; Fouhse, J.; Im, H.S.; Chan, C.B.; Willing, B.P. Dietary Pea Fiber Supplementation Improves Glycemia and Induces Changes in the Composition of Gut Microbiota, Serum Short Chain Fatty Acid Profile and Expression of Mucins in Glucose Intolerant Rats. Nutrients 2017, 9, 1236. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Li, Q.; Hu, K.; He, Y.; Ai, Q.; Hu, L.; Yu, J. Vitamin A and Retinoic Acid Exhibit Protective Effects on Necrotizing Enterocolitis by Regulating Intestinal Flora and Enhancing the Intestinal Epithelial Barrier. Arch. Med. Res. 2018, 49, 1–9. [Google Scholar] [CrossRef]
- Luthold, R.V.; Fernandes, G.R.; Franco-De-Moraes, A.C.; Folchetti, L.G.; Ferreira, S.R.G. Gut microbiota interactions with the immunomodulatory role of vitamin D in normal individuals. Metabolism 2017, 69, 76–86. [Google Scholar] [CrossRef]
- Sun, H.; Chen, Y.; Cheng, M.; Zhang, X.; Zheng, X.; Zhang, Z. The modulatory effect of polyphenols from green tea, oolong tea and black tea on human intestinal microbiota in vitro. J. Food Sci. Technol. 2018, 55, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Camps-Bossacoma, M.; Pérez-Cano, F.J.; Franch, A.; Castell, M. Gut Microbiota in a Rat Oral Sensitization Model: Effect of a Cocoa-Enriched Diet. Oxidative Med. Cell. Longev. 2017, 2017, 1–12. [Google Scholar] [CrossRef]
- Gowd, V.; Karim, N.; Shishir, M.R.I.; Xie, L.; Chen, W. Dietary polyphenols to combat the metabolic diseases via altering gut microbiota. Trends Food Sci. Technol. 2019, 93, 81–93. [Google Scholar] [CrossRef]
- Christmann, B.S.; Abrahamsson, T.R.; Bernstein, C.N.; Duck, L.W.; Mannon, P.J.; Berg, G.; Björkstén, B.; Jenmalm, M.C.; Elson, C.O. Human seroreactivity to gut microbiota antigens. J. Allergy Clin. Immunol. 2015, 136, 1378–1386.e5. [Google Scholar] [CrossRef]
- Azad, M.B.; Konya, T.; Guttman, D.S.; Field, C.; Sears, M.R.; HayGlass, K.T.; Mandhane, P.J.; Turvey, S.; Subbarao, P.; Becker, A.B.; et al. Infant gut microbiota and food sensitization: Associations in the first year of life. Clin. Exp. Allergy 2015, 45, 632–643. [Google Scholar] [CrossRef]
- Qin, Y.; Wang, M.; Dong, P. Association Between Food Allergy and Microbiome Status in 1-Year-Old Infants in STRONG Kids 2 Cohort; Illinois Library: Urbana, IL, USA, 2018; Available online: https://hdl.handle.net/2142/99865 (accessed on 1 April 2018).
- Ling, Z.; Li, Z.; Liu, X.; Cheng, Y.; Luo, Y.; Tong, X.; Yuan, L.; Wang, Y.; Sun, J.; Li, L.; et al. Altered Fecal Microbiota Composition Associated with Food Allergy in Infants. Appl. Environ. Microbiol. 2014, 80, 2546–2554. [Google Scholar] [CrossRef]
- Chen, C.; Chen, K.-J.; Kong, M.; Chang, H.; Huang, J. Alterations in the gut microbiotas of children with food sensitization in early life. Pediatr. Allergy Immunol. 2016, 27, 254–262. [Google Scholar] [CrossRef]
- Inoue, R.; Sawai, T.; Sawai, C.; Nakatani, M.; Romero-Pérez, G.A.; Ozeki, M.; Nonomura, K.; Tsukahara, T. A preliminary study of gut dysbiosis in children with food allergy. Biosci. Biotechnol. Biochem. 2017, 81, 2396–2399. [Google Scholar] [CrossRef]
- Lee, K.; Guo, J.; Song, Y.; Ariff, A.; O’Sullivan, M.; Hales, B.; Mullins, B.; Zhang, G. Dysfunctional Gut Microbiome Networks in Childhood IgE-Mediated Food Allergy. Int. J. Mol. Sci. 2021, 22, 2079. [Google Scholar] [CrossRef]
- Fazlollahi, M.; Chun, Y.; Grishin, A.; Wood, R.A.; Burks, A.W.; Dawson, P.; Jones, S.M.; Leung, D.Y.M.; Sampson, H.A.; Sicherer, S.H.; et al. Early-life gut microbiome and egg allergy. Allergy 2018, 73, 1515–1524. [Google Scholar] [CrossRef]
- Bunyavanich, S.; Shen, N.; Grishin, A.; Wood, R.; Burks, W.; Dawson, P.; Jones, S.M.; Leung, D.Y.; Sampson, H.; Sicherer, S.; et al. Early-life gut microbiome composition and milk allergy resolution. J. Allergy Clin. Immunol. 2016, 138, 1122–1130. [Google Scholar] [CrossRef]
- Hua, X.; Goedert, J.J.; Pu, A.; Yu, G.; Shi, J. Allergy associations with the adult fecal microbiota: Analysis of the American Gut Project. eBioMedicine 2016, 3, 172–179. [Google Scholar] [CrossRef]
- Liu, Q.; Jing, W.; Wang, W. Bifidobacterium lactis Ameliorates the Risk of Food Allergy in Chinese Children by Affecting Relative Percentage of Treg and Th17 Cells. Can. J. Infect. Dis. Med. Microbiol. 2018, 2018, 4561038. [Google Scholar] [CrossRef]
- Savage, J.H.; Lee-Sarwar, K.A.; Sordillo, J.; Bunyavanich, S.; Zhou, Y.; O’Connor, G.; Sandel, M.; Bacharier, L.B.; Zeiger, R.; Sodergren, E.; et al. A prospective microbiome-wide association study of food sensitization and food allergy in early childhood. Allergy 2018, 73, 145–152. [Google Scholar] [CrossRef]
- Wu, W.; Sun, M.; Chen, F.; Cao, A.T.; Liu, H.; Zhao, Y.; Huang, X.; Xiao, Y.; Yao, S.; Zhao, Q.; et al. Microbiota metabolite short-chain fatty acid acetate promotes intestinal IgA response to microbiota which is mediated by GPR43. Mucosal Immunol. 2017, 10, 946–956. [Google Scholar] [CrossRef]
- Trompette, A.; Gollwitzer, E.S.; Yadava, K.; Sichelstiel, A.K.; Sprenger, N.; Ngom-Bru, C.; Blanchard, C.; Junt, T.; Nicod, L.P.; Harris, N.L.; et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014, 20, 159–166. [Google Scholar] [CrossRef]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, Receptor for Niacin and the Commensal Metabolite Butyrate, Suppresses Colonic Inflammation and Carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef]
- Park, J.; Kim, M.; Kang, S.G.; Jannasch, A.H.; Cooper, B.; Patterson, J.; Kim, C.H. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR–S6K pathway. Mucosal Immunol. 2015, 8, 80–93. [Google Scholar] [CrossRef]
- Michaudel, C.; Sokol, H. The Gut Microbiota at the Service of Immunometabolism. Cell Metab. 2020, 32, 514–523. [Google Scholar] [CrossRef]
- Thorburn, A.N.; McKenzie, C.I.; Shen, S.; Stanley, D.; Macia, L.; Mason, L.J.; Roberts, L.K.; Wong, C.H.Y.; Shim, R.; Robert, R.; et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat. Commun. 2015, 6, 7320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macia, L.; Tan, J.; Vieira, A.T.; Leach, K.; Stanley, D.; Luong, S.; Maruya, M.; McKenzie, C.l.; Hijikata, A.; Wong, C.; et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat. Commun. 2015, 6, 6734. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Wang, Y.; Wang, P.; Huang, Y.; Wang, F. Short-Chain Fatty Acids Manifest Stimulative and Protective Effects on Intestinal Barrier Function Through the Inhibition of NLRP3 Inflammasome and Autophagy. Cell. Physiol. Biochem. 2018, 49, 190–205. [Google Scholar] [CrossRef] [PubMed]
- Kepert, I.; Fonseca, J.; Müller, C.; Milger, K.; Hochwind, K.; Kostric, M.; Fedoseeva, M.; Ohnmacht, C.; Dehmel, S.; Nathan, P.; et al. D-tryptophan from probiotic bacteria influences the gut microbiome and allergic airway disease. J. Allergy Clin. Immunol. 2017, 139, 1525–1535. [Google Scholar] [CrossRef]
- Crestani, E.; Harb, H.; Charbonnier, L.-M.; Leirer, J.; Motsinger-Reif, A.; Rachid, R.; Phipatanakul, W.; Kaddurah-Daouk, R.; Chatila, T.A. Untargeted metabolomic profiling identifies disease-specific signatures in food allergy and asthma. J. Allergy Clin. Immunol. 2020, 145, 897–906. [Google Scholar] [CrossRef]
- Venkatesh, M.; Mukherjee, S.; Wang, H.; Li, H.; Sun, K.; Benechet, A.P.; Qiu, Z.; Maher, L.; Redinbo, M.R.; Phillips, R.S.; et al. Symbiotic Bacterial Metabolites Regulate Gastrointestinal Barrier Function via the Xenobiotic Sensor PXR and Toll-like Receptor 4. Immunity 2014, 41, 296–310. [Google Scholar] [CrossRef]
- Song, X.; Sun, X.; Oh, S.F.; Wu, M.; Zhang, Y.; Zheng, W.; Geva-Zatorsky, N.; Jupp, R.; Mathis, D.; Benoist, C.; et al. Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis. Nature 2020, 577, 410–415. [Google Scholar] [CrossRef]
- Quraishi, M.N.; Shaheen, W.; Oo, Y.H.; Iqbal, T.H. Immunological mechanisms underpinning faecal microbiota transplantation for the treatment of inflammatory bowel disease. Clin. Exp. Immunol. 2020, 199, 24–38. [Google Scholar] [CrossRef]
- Feehley, T.; Nagler, C.R. Cellular and molecular pathways through which commensal bacteria modulate sensitization to dietary antigens. Curr. Opin. Immunol. 2014, 31, 79–86. [Google Scholar] [CrossRef]
- Kulkarni, D.H.; Gustafsson, J.K.; Knoop, K.A.; McDonald, K.G.; Bidani, S.S.; Davis, J.E.; Floyd, A.N.; Hogan, S.P.; Hsieh, C.-S.; Newberry, R.D. Goblet cell associated antigen passages support the induction and maintenance of oral tolerance. Mucosal Immunol. 2020, 13, 271–282. [Google Scholar] [CrossRef]
- Stefka, A.T.; Feehley, T.; Tripathi, P.; Qiu, J.; McCoy, K.; Mazmanian, S.K.; Tjota, M.Y.; Seo, G.-Y.; Cao, S.; Theriault, B.R.; et al. Commensal bacteria protect against food allergen sensitization. Proc. Natl. Acad. Sci. USA 2014, 111, 13145–13150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sardecka-Milewska, I.; Łoś-Rycharska, E.; Gawryjołek, J.; Kowalska, E.T.; Krogulska, A. Role of FOXP3 Expression and Serum Vitamin D and C Concentrations When Predicting Acquisition of Tolerance in Infants With Cow’s Milk Allergy. J. Investig. Allergy Clin. Immunol. 2019, 30, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; McKenzie, C.; Vuillermin, P.J.; Goverse, G.; Vinuesa, C.G.; Mebius, R.E.; Macia, L.; Mackay, C.R. Dietary Fiber and Bacterial SCFA Enhance Oral Tolerance and Protect against Food Allergy through Diverse Cellular Pathways. Cell Rep. 2016, 15, 2809–2824. [Google Scholar] [CrossRef] [PubMed]
- Michels, K.R.; Lukacs, N.W.; Fonseca, W. TLR Activation and Allergic Disease: Early Life Microbiome and Treatment. Curr. Allergy Asthma Rep. 2018, 18, 61. [Google Scholar] [CrossRef]
- Pernomian, L.; Duarte-Silva, M.; Cardoso, C.R.D.B. The Aryl Hydrocarbon Receptor (AHR) as a Potential Target for the Control of Intestinal Inflammation: Insights from an Immune and Bacteria Sensor Receptor. Clin. Rev. Allergy Immunol. 2020, 59, 382–390. [Google Scholar] [CrossRef]
- Caminero, A.; Meisel, M.; Jabri, B.; Verdu, E.F. Mechanisms by which gut microorganisms influence food sensitivities. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 7–18. [Google Scholar] [CrossRef]
- Al-Sahlany, S.T.; Niamah, A.K. Bacterial viability, antioxidant stability, antimutagenicity and sensory properties of onion types fermentation by using probiotic starter during storage. Nutr. Food Sci. 2022, 52, 901–916. [Google Scholar] [CrossRef]
- Berni Canani, R.; Di Costanzo, M.; Bedogni, G.; Amoroso, A.; Cosenza, L.; Di Scala, C.; Granata, V.; Nocerino, R. Extensively hydrolyzed casein formula containing Lactobacillus rhamnosus GG reduces the occurrence of other allergic manifestations in children with cow’s milk allergy: 3-year randomized controlled trial. J. Allergy Clin. Immunol. 2017, 139, 1906–1913.e4. [Google Scholar] [CrossRef]
- Berni Canani, R.; Sangwan, N.; Stefka, A.; Nocerino, R.; Paparo, L.; Aitoro, R.; Calignano, A.; Khan, A.A.; Gilbert, J.A.; Nagler, C.R. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J. 2016, 10, 742–750. [Google Scholar] [CrossRef]
- Bu, G.; Luo, Y.; Zhang, Y.; Chen, F. Effects of fermentation by lactic acid bacteria on the antigenicity of bovine whey proteins. J. Sci. Food Agric. 2010, 90, 2015–2020. [Google Scholar] [CrossRef]
- Bertrand-Harb, C.; Ivanova, I.; Dalgalarrondo, M.; Haertllé, T. Evolution of β-lactoglobulin and α-lactalbumin content during yoghurt fermentation. Int. Dairy J. 2003, 13, 39–45. [Google Scholar] [CrossRef]
- Rizzello, C.G.; De Angelis, M.; Coda, R.; Gobbetti, M. Use of selected sourdough lactic acid bacteria to hydrolyze wheat and rye proteins responsible for cereal allergy. Eur. Food Res. Technol. 2006, 223, 405–411. [Google Scholar] [CrossRef]
- De Angelis, M.; Rizzello, C.G.; Scala, E.; De Simone, C.; Farris, G.A.; Turrini, F.; Gobbetti, M. Probiotic Preparation Has the Capacity To Hydrolyze Proteins Responsible for Wheat Allergy. J. Food Prot. 2007, 70, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Xiao, L.; Liu, S.; Liu, X.; Luo, Y.; Ji, Q.; Yang, P.; Liu, Z. Exploration of the effect of probiotics supplementation on intestinal microbiota of food allergic mice. Am. J. Transl. Res. 2017, 9, 376–385. [Google Scholar] [PubMed]
- Fu, L.; Song, J.; Wang, C.; Fu, S.; Wang, Y. Bifidobacterium infantis Potentially Alleviates Shrimp Tropomyosin-Induced Allergy by Tolerogenic Dendritic Cell-Dependent Induction of Regulatory T Cells and Alterations in Gut Microbiota. Front. Immunol. 2017, 8, 1536. [Google Scholar] [CrossRef] [PubMed]
- Neau, E.; Delannoy, J.; Marion, C.; Cottart, C.-H.; Labellie, C.; Holowacz, S.; Butel, M.-J.; Kapel, N.; Waligora-Dupriet, A.-J. Three Novel Candidate Probiotic Strains with Prophylactic Properties in a Murine Model of Cow’s Milk Allergy. Appl. Environ. Microbiol. 2016, 82, 1722–1733. [Google Scholar] [CrossRef] [PubMed]
- Kozakova, H.; Schwarzer, M.; Srutkova, D.; Hudcovic, T.; Cukrowska, B. Colonisation of germ-free mice with probiotic lactobacilli mitigated allergic sensitisation in murine model of birch pollen allergy. Clin. Transl. Allergy 2014, 4, P26. [Google Scholar] [CrossRef]
- Fang, S.; Zhang, Y.; Zhang, Y.; Zhu, X.; Yie, B. Lactobacillus rhamnosus GG improves symptoms and its mechnism in mice with ovalbumin-induced food allergy. Chin. J. Cell. Mol. Immunol. 2017, 33, 597–600. [Google Scholar]
- Shandilya, U.K.; Sharma, A.; Kapila, R.; Kansal, V.K. Probiotic Dahi containing Lactobacillus acidophilus and Bifidobacterium bifidum modulates immunoglobulin levels and cytokines expression in whey proteins sensitised mice. J. Sci. Food Agric. 2016, 96, 3180–3187. [Google Scholar] [CrossRef]
- Li, N.; Yu, Y.; Chen, X.; Gao, S.; Zhang, Q.; Xu, C. Bifidobacterium breve M-16V alters the gut microbiota to alleviate OVA-induced food allergy through IL-33/ST2 signal pathway. J. Cell. Physiol. 2020, 235, 9464–9473. [Google Scholar] [CrossRef]
- Cortes-Perez, N.; Lozano-Ojalvo, D.; Maiga, M.; Hazebrouck, S.; Adel-Patient, K. Intragastric administration of Lactobacillus casei BL23 induces regulatory FoxP3+RORγt+ T cells subset in mice. Benef. Microbes 2017, 8, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Suarez, P.; Gueimonde, M.; Margolles, A.; Suarez-Diaz, A.M. Distinct Bifidobacterium strains drive different immune responses in vitro. Int. J. Food Microbiol. 2010, 138, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Price, D.B.; Ackland, M.L.; Burks, W.; Knight, M.I.; Suphioglu, C. Peanut Allergens Alter Intestinal Barrier Permeability and Tight Junction Localisation in Caco-2 Cell Cultures. Cell. Physiol. Biochem. 2014, 33, 1758–1777. [Google Scholar] [CrossRef] [PubMed]
- Chichlowski, M.; De Lartigue, G.; German, J.B.; Raybould, H.E.; Mills, D.A. Bifidobacteria Isolated From Infants and Cultured on Human Milk Oligosaccharides Affect Intestinal Epithelial Function. J. Pediatr. Gastroenterol. Nutr. 2012, 55, 321–327. [Google Scholar] [CrossRef] [PubMed]
- de Kivit, S.; Tobin, M.C.; DeMeo, M.T.; Fox, S.; Garssen, J.; Forsyth, C.B.; Keshavarzian, A.; Landay, A.L. In vitro evaluation of intestinal epithelial TLR activation in preventing food allergic responses. Clin. Immunol. 2014, 154, 91–99. [Google Scholar] [CrossRef]
- Wu, Z.; Nasab, E.M.; Arora, P.; Athari, S.S. Study effect of probiotics and prebiotics on treatment of OVA-LPS-induced of allergic asthma inflammation and pneumonia by regulating the TLR4/NF-kB signaling pathway. J. Transl. Med. 2022, 20, 130. [Google Scholar] [CrossRef]
- Capurso, L. Thirty Years of Lactobacillus rhamnosus GG. J. Clin. Gastroenterol. 2019, 53, S1–S41. [Google Scholar] [CrossRef]
- Jerzynska, J.; Stelmach, W.; Balcerak, J.; Woicka-Kolejwa, K.; Rychlik, B.; Blauz, A.; Wachulec, M.; Stelmach, P.; Majak, P.; Stelmach, I. Effect of Lactobacillus rhamnosus GG and vitamin D supplementation on the immunologic effectiveness of grass-specific sublingual immunotherapy in children with allergy. Allergy Asthma Proc. 2016, 37, 324–334. [Google Scholar] [CrossRef]
- Ouwehand, A.C.; Nermes, M.; Collado, M.C.; Rautonen, N.; Salminen, S.; Isolauri, E. Specific probiotics alleviate allergic rhinitis during the birch pollen season. World J. Gastroenterol. 2009, 15, 3261–3268. [Google Scholar] [CrossRef]
- Plummer, E.L.; Lozinsky, A.C.; Tobin, J.M.; Uebergang, J.B.; Axelrad, C.; Garland, S.M.; Jacobs, S.E.; Tang, M.L.K.; ProPrems Study Group. Postnatal probiotics and allergic disease in very preterm infants: Sub-study to the ProPrems randomized trial. Allergy 2020, 75, 127–136. [Google Scholar] [CrossRef]
- West, C.E.; Hammarström, M.-L.; Hernell, O. Probiotics in primary prevention of allergic disease—Follow-up at 8–9 years of age. Allergy 2013, 68, 1015–1020. [Google Scholar] [CrossRef] [PubMed]


| Allergens Involved | Study Population | Age | Changes of Gut Microbiota in Food Allergy | Reference |
|---|---|---|---|---|
| Milk, eggs, wheat, nut, peanuts, fish, shrimp, soybeans | 166 infants | 0 to 1 year | ↑ Enterobacteriaceae/Bacteroidaceae ratio ↓ Ruminococcaceae | [36] |
| Milk, eggs, wheat, nut, peanuts, fish, shrimp, soybeans | 49 healthy infants 38 infants with food allergy | 1 week to 12 months of age | ↑ Bifidobacterium spp. — Lactobacillus spp. and Clostridium perfringens | [37] |
| Milk, eggs, wheat, nut, peanuts, fish, shrimp, soybean | 34 infants with food allergy 45 healthy controls | 2 to 11 years | ↑ Firmicutes and Fusobacteria ↓ Bacteroidetes, Proteobacteria and Actinobacteria ↑ Clostridium sensustricto and Anaerobacter ↓ Bacteroides and Clostridium XVIII | [38] |
| Egg white, cow’s milk, wheat, peanut, soybean, gluten | 23 children with food allergy 22 healthy children | 6 to 24 months | ↑ Sphingomonas, Sutterella, Bifidobacterium, Collinsella, Clostridium sensustricto, Clostridium IV, Enterococcus, Lactobacillus, Roseburia, Faecalibacterium, Ruminococcus, Subdoligranulum, and Akkermansia, ↓ Bacteroides, Parabacteroides, Prevotella, Alistipes, Streptococcus, and Veillonella | [39] |
| Egg, soybean, sesame, milk, shrimp, crab, peanut, wheat | 4 children with food allergy 4 healthy children | 18 months to 6 years | ↓ Dorea and Akkermansia ↑ Lachnospira, Veillonella, and Sutterella | [40] |
| Egg | 141 children with egg allergy | 3 to 16 months | Lachnospiraceae, Streptococcaceae, and Leuconostocaceae families were differentially abundant in children with egg allergy | [42] |
| Cow milk | 226 children with milk allergy | 3 to 16 months | Clostridia and Firmicutes could be studied as probiotic candidates for milk allergy therapy | [43] |
| Peanuts, tree nuts, shellfish | 1879 participants was 81.5%, ranging from 2.5% for peanuts to 40.5% for seasonal. | mean age, 45.5 years; 46.9% male | higher Bacteroidales and reduced Clostridiales taxa in nut and seasonal allergies | [44] |
| Egg, crab, shrimp | 256 children with food allergy | 4 to 12 years | Bifidobacterium lactis can effectively alleviate allergic reactions on food-specific IgE of food in children | [45] |
| SCFAs | Pathway | Proposed Mechanism | Reference |
|---|---|---|---|
| Acetate | GPR43 | Promote gut IgA responses | [47] |
| Propionate | GPR41 | Damage Th2 effector cells | [48] |
| Butyrate | GPR109a | Induction of Treg cells and IL-18 in the immune tolerance of food allergens | [49] |
| Acetate, propionate, butyrate | HDAC | Promote the differentiation of T cells into Th1, Th17 and Treg cells | [50] |
| Valerate | HDAC | Increase the production of anti-inflammatory factor IL-10 | [51] |
| Acetate | HDAC9 | Accelerate the production of Treg cells to alleviate allergic diseases | [52] |
| Subject | Source of Probiotics | Allergens Source | Therapies and Supplements | Effects and Mechanism of Action | Reference |
|---|---|---|---|---|---|
| human | Lacticaseibacillus rhamnosus GG (LGG) | Cow milk | Children with IgE-mediated cow milk allergy were randomly allocated to the LGG and followed for 36 months. | Reduced the incidence of the other allergic manifestations and hastens the development of oral tolerance in children with IgE-mediated cow’s milk allergy by a favorable modulation of gut microbiota and epigenetic mechanisms. | [69] |
| LGG | Cow milk | Infant with IgE-mediated cow milk allergy were treated with extensively hydrolyzed casein formula either with or without supplementation with LGG (at 4.5 × 107–8.5 × 107 CFU/g) for 6 months. | Promoted tolerance in infants with cow’s milk allergy by influencing the strain-level bacterial community structure of the infant gut. | [70] | |
| Lactobacillus helveticus and Streptococcus thermophilus | α-lactalbumin and β-lactoglobul | Fermentation | The fermentation with lactic acid bacteria is an effective way to reduce whey proteins antigenicity. | [71] | |
| Streptococcus thermophilus and Lactobacillus delbrueckii subsp. bulgaricus | α-lactalbumin and β-lactoglobul | Lactobacillus fermented milk can not only interrupt allergen epitopes and produce some bioactive peptides, but also regulate the immune system. | [72] | ||
| Lactobacillus alimentarius, Lactobacillus brevis, Lactobacillus sanfranciscensis and Lactobacillus hilgardii | Wheat allergens | Lactic acid bacteria fermentation was more conducive to the degradation of IgE epitopes of wheat protein. | [73] | ||
| Probiotic VSL#3 (Streptococcus thermophilus, Lactobacillus plantarum, L. acidophilus, L. casei, L. delbrueckii spp. bulgaricus, Bifidobacterium breve, B. longum and B. infantis) | Wheat allergens | Probiotic VSL#3 can reduce the sensitizing proteins that cannot be degraded by pepsin and trypsin and reduce the potential risk of wheat protein to a certain extent. | [74] | ||
| animal | Bifidobacterium | Ovalbumin | Mice were orally administered with 200 mL/mouse of normal saline containing 108 CFU/mL for 2 weeks. | Probiotics ameliorated allergic symptoms, including reducing OVA specific-IgE, and -IgG1 levels in the serum, Th2 cytokines release in spleen, and occurrence of diarrhea. Moreover, 16S rRNA analysis showed that the probiotics-mediated protection was conferred by an enrichment of Coprococcus and Rikenella. | [75] |
| Bifidobacterium infantis 14.518 (Binf) | Shrimp-tropomyosin (TM) | Mice were daily administered with 500 µL/mouse Binf (2 × 109 CFU/mL) for 3 weeks | Binf promotes the induction of Tregs and balance Th2/Treg for suppressing Th2 responses in TM-sensitized mice. | [76] | |
| Lacticaseibacillus rhamnosus, Lactobacillus salivarius, and Bifidobacterium longum subsp. infantis | Cow milk | 109 CFU/g for 6 weeks in mice | Lactobacillus salivarius strain blocked Th1 and Th2 responses, while the Bifidobacterium longum subsp. infantis strain induced a pro-Th1 profile and the Lacticaseibacillus rhamnosus strain induced pro-Th1 and regulatory response. | [77] | |
| animal | Lacticaseibacillus rhamnosus and Lacticaseibacillus casei | Birch pollen | HEK293 cells of mice were stimulated with the formalin-inactivated Lacticaseibacillus rhamnosus and Lacticaseibacillus casei or their equal-part mixture at a concentration of 107 CFU/mL for 20 h | Colonization with lactobacilli mixture inhibited the development of allergic immune responses through induction of regulatory cytokine TGF-β and can be thus exploited for alleviation of pollen allergies. | [78] |
| LGG | Ovalbumin | Mice were given LGG for consecutive 22 days (low-dose LGG group, 1 × 108 CFU/mL, 200 μL/d and high-dose LGG group, 1 × 109 CFU/mL, 200 μL/d | Lacticaseibacillus rhamnosus GG can remarkably improve the symptoms of ovalbumin-induced food allergy probably by decreasing IL-4/IFN-γ ratio. | [79] | |
| La-Dahi (Lactobacillus acidophilus LaVK2 and Bifidobacterium bifidum BbVK3) | Whey proteins | 2 × 109 CFU/g for 1 week in mice | Probiotic Dahi skewed Th2-specific immune response towards Th1-specific response and suppressed IgE in serum | [80] | |
| Bifidobacterium breve M-16V | Ovalbumin | 1 × 107 CFU/g formula per mouse for 40 days | Bifidobacterium breve M-16V may alter the gut microbiota to alleviate the allergy symptoms by IL-33/tumorigenicity 2 signaling | [81] | |
| Lacticaseibacillus casei BL23 | Cow milk | Mice were intragastrically administered with Lacticaseibacillus casei BL23 (~1.5 × 108 CFU/mouse/administration) for 5 consecutive days. | Intragastric administration of Lacticaseibacillus casei BL23 to mice induces local and systemic Foxp3+ RORγt+ type 3 Treg cells, the specific transcription factor of Th17 cells. | [82] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, H.; Qu, Y.; Gao, Y.; Sun, S.; Wu, R.; Wu, J. Research Progress on the Correlation between the Intestinal Microbiota and Food Allergy. Foods 2022, 11, 2913. https://doi.org/10.3390/foods11182913
Yang H, Qu Y, Gao Y, Sun S, Wu R, Wu J. Research Progress on the Correlation between the Intestinal Microbiota and Food Allergy. Foods. 2022; 11(18):2913. https://doi.org/10.3390/foods11182913
Chicago/Turabian StyleYang, Hui, Yezhi Qu, Yaran Gao, Shuyuan Sun, Rina Wu, and Junrui Wu. 2022. "Research Progress on the Correlation between the Intestinal Microbiota and Food Allergy" Foods 11, no. 18: 2913. https://doi.org/10.3390/foods11182913
APA StyleYang, H., Qu, Y., Gao, Y., Sun, S., Wu, R., & Wu, J. (2022). Research Progress on the Correlation between the Intestinal Microbiota and Food Allergy. Foods, 11(18), 2913. https://doi.org/10.3390/foods11182913