Characterization, High-Density Fermentation, and the Production of a Directed Vat Set Starter of Lactobacilli Used in the Food Industry: A Review
Abstract
:1. Introduction
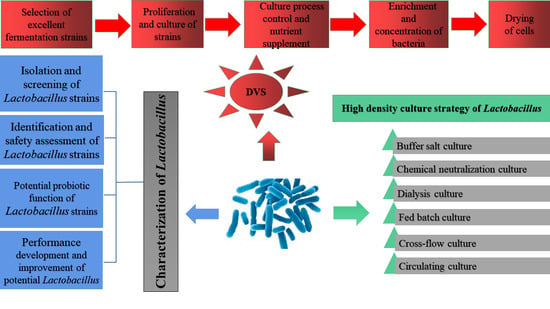
2. Characterization of Lactobacilli Strains
2.1. Screening out Lactobacilli Strains
2.2. Identification and Safety Assessment of Lactobacilli Strains
2.3. Potential Probiotic Functionalities of Lactobacilli Strains
2.4. Fermentation Performance of Lactobacilli Strains
2.5. Health Functions of Lactobacilli Strains
2.6. Performance Development and Improvement of Lactobacilli Strains
2.7. Role of Lactobacilli Strains in Food Production
3. High-Density Fermentation
| Current High-Density-Culture Methods | Advantage | Disadvantages |
|---|---|---|
| Buffer salt culture [179] | Add a buffer salt that has no effect on the strain or has a growth-promoting impact on the culture medium to improve the buffering capacity of the fermentation broth and control the stability of pH within a specific range, easy to operate. | The buffering capacity of the buffer salt is limited and can only play a role within a specific range. |
| Chemical neutralization culture [180] | Add lye (such as NaOH, ammonia, and CaCO3) to the culture system to control the pH value of the fermentation system, easy to operate. | With the continuous addition of lye and the accumulation of metabolites, too high salt concentration will inhibit the growth of bacteria. |
| Dialysis culture [181] | Remove part of the small molecular metabolites produced by the bacteria while providing fresh nutrients to the culture solution. | A small processing volume, a long dialysis process, and large equipment investment are also not conducive to industrialization. |
| Fed-batch culture (non-feedback mode and feedback mode) [182,183,184,185] | Effectively eliminates substrate inhibition and acid inhibition and is simple to operate | Inadequate utilization of nutrients; Limited by container volume |
| Cross-flow culture [186] | Due to cross-flow filtration, the high viscosity produced by cells is reduced, which is conducive to cell recovery and high concentration | High equipment cost; Requires more professional operators; It is easy to block the membrane module. |
| Circulating culture (sedimentation, centrifugation, and membrane filtration) [187,188] | Through technologies such as sedimentation, centrifugation, and membrane filtration, the cells are intercepted, the culture medium flows out, and then a certain amount of fresh culture medium is added to obtain high-density cells. It shortens the production time and saves a lot of power, workforce, water, and steam | In the circulation process, the strains quickly degenerate and are polluted, resulting in economic losses; The utilization rate of nutrients was lower than that of batch culture. |
4. Production of DVS Starter of Lactobacilli
5. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Iseppi, R.; Zurlini, C.; Cigognini, I.M.; Cannavacciuolo, M.; Sabia, C.; Messi, P. Eco-friendly edible packaging systems based on live-Lactobacillus kefiri MM5 for the control of Listeria monocytogenes in fresh vegetables. Foods 2022, 11, 2632. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wang, Y.; Han, Y.; Zhang, J.; Wang, S.; Lu, S.; Wang, H.; Lu, F.; Jia, L. Complete genome sequence of a novel Lactobacillus paracasei TK1501 and its application in the biosynthesis of isoflavone aglycones. Foods 2022, 11, 2807. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; Toole, P.W.O.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Micr. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Reid, G. Probiotics: Definition, scope and mechanisms of action. Best Pract. Res. Clin. Gastroenterol. 2016, 30, 17–25. [Google Scholar] [CrossRef]
- Brizuela, N.; Tymczyszyn, E.E.; Semorile, L.C.; Valdes La Hens, D.; Delfederico, L.; Hollmann, A.; Bravo-Ferrada, B. Lactobacillus plantarum as a malolactic starter culture in winemaking: A new (old) player? Electron. J. Biotechn. 2019, 38, 10–18. [Google Scholar] [CrossRef]
- Shori, A.B.; Aljohani, G.S.; Al-zahrani, A.J.; Al-sulbi, O.S.; Baba, A.S. Viability of probiotics and antioxidant activity of cashew milk-based yogurt fermented with selected strains of probiotic Lactobacillus spp. LWT 2022, 153, 112482. [Google Scholar] [CrossRef]
- Yang, W.; Hao, X.; Zhang, X.; Zhang, G.; Li, X.; Liu, L.; Sun, Y.; Pan, Y. Identification of antioxidant peptides from cheddar cheese made with Lactobacillus helveticus. LWT 2021, 141, 110866. [Google Scholar] [CrossRef]
- Ilha, E.C.; Da Silva, T.; Lorenz, J.G.; de Oliveira Rocha, G.; Sant Anna, E.S. Lactobacillus paracasei isolated from grape sourdough: Acid, bile, salt, and heat tolerance after spray drying with skim milk and cheese whey. Eur. Food Res. Technol. 2015, 240, 977–984. [Google Scholar] [CrossRef]
- Sun, F.; Kong, B.; Chen, Q.; Han, Q.; Diao, X. N-nitrosoamine inhibition and quality preservation of Harbin dry sausages by inoculated with Lactobacillus pentosus, Lactobacillus curvatus and Lactobacillus sake. Food Control 2017, 73, 1514–1521. [Google Scholar] [CrossRef]
- Zhou, M.; Zheng, X.; Zhu, H.; Li, L.; Zhang, L.; Liu, M.; Liu, Z.; Peng, M.; Wang, C.; Li, Q.; et al. Effect of Lactobacillus plantarum enriched with organic/inorganic selenium on the quality and microbial communities of fermented pickles. Food Chem. 2021, 365, 130495. [Google Scholar] [CrossRef]
- Guantario, B.; Zinno, P.; Schifano, E.; Roselli, M.; Perozz, G.; Uccelletti, C.P.D.; Devirgiliis, C. In vitro and in vivo selection of potentially probiotic Lactobacilli from Nocellara del Belice table olives. Front. Microbiol. 2018, 9, 595. [Google Scholar] [CrossRef] [Green Version]
- Song, J.; Peng, S.; Yang, J.; Zhou, F.; Suo, H. Isolation and identification of novel antibacterial peptides produced by Lactobacillus fermentum SHY10 in Chinese pickles. Food Chem. 2021, 348, 129097. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zheng, J.; He, L.; Li, C.; Hu, P.; Tao, H.; Wang, X. Evaluation of the effect of essential oil addition on the quality parameters and predicted shelf life of potato yogurt. J. Food Protect. 2021, 84, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- Shori, A.B.; Rashid, F.; Baba, A.S. Effect of the addition of phytomix-3+ mangosteen on antioxidant activity, viability of lactic acid bacteria, type 2 diabetes key-enzymes, and sensory evaluation of yogurt. LWT 2018, 94, 33–39. [Google Scholar] [CrossRef]
- Divyashree, S.; Anjali, P.G.; Somashekaraiah, R.; Sreenivasa, M.Y. Probiotic properties of Lactobacillus casei—MYSRD 108 and Lactobacillus plantarum-MYSRD 71 with potential antimicrobial activity against Salmonella paratyphi. Biotechnol. Rep. 2021, 32, e00672. [Google Scholar] [CrossRef]
- Xu, J.; Guo, L.; Zhao, N.; Meng, X.; Zhang, J.; Wang, T.; Wei, X.; Fan, M. Response mechanisms to acid stress of acid-resistant bacteria and biotechnological applications in the food industry. Crit. Rev. Biotechnol. 2022, 42, 1–17. [Google Scholar] [CrossRef]
- García-Gamboa, R.G.M.O. Assessment of intermediate and long chains agave fructan fermentation on the growth of intestinal bacteria cultured in a gastrointestinal tract simulator. Rev. Mex. Ing. Quim. 2019, 19, 827–838. [Google Scholar] [CrossRef] [Green Version]
- Tuo, Y.; Yu, H.; Ai, L.; Wu, Z.; Guo, B.; Chen, W. Aggregation and adhesion properties of 22 Lactobacillus strains. J. Dairy Sci. 2013, 96, 4252–4257. [Google Scholar] [CrossRef] [Green Version]
- Gaspar, C.; Donders, G.G.; de Oliveira, R.P.; Queiroz, J.A.; Tomaz, C.; de Oliveira, J.M.; de Oliveira, A.P. Bacteriocin production of the probiotic Lactobacillus acidophilus KS400. AMB Express 2018, 8, 153. [Google Scholar] [CrossRef]
- Toropov, V.; Demyanova, E.; Shalaeva, O.; Sitkin, S.; Vakhitov, T. Whole-genome sequencing of Lactobacillus helveticus D75 and D76 confirms safety and probiotic potential. Microorganisms 2020, 8, 329. [Google Scholar] [CrossRef]
- Meireles Mafaldo, Í.; de Medeiros, V.P.B.; Da Costa, W.K.A.; Da Costa Sassi, C.F.; Da Costa Lima, M.; de Souza, E.L.; Eduardo Barão, C.; Colombo Pimentel, T.; Magnani, M. Survival during long-term storage, membrane integrity, and ultrastructural aspects of Lactobacillus acidophilus 05 and Lacticaseibacillus casei 01 freeze-dried with freshwater microalgae biomasses. Food Res. Int. 2022, 159, 111620. [Google Scholar] [CrossRef] [PubMed]
- Whittington, H.D.; Dagher, S.F.; Bruno-Bárcena, J.M. Production and conservation of starter cultures: From “backslopping” to controlled fermentations. In How Fermented Foods Feed a Healthy Gut Microbiota: A Nutrition Continuum; Azcarate-Peril, M.A., Arnold, R.R., Bruno-Bárcena, J.M., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 125–138. [Google Scholar] [CrossRef]
- Reque, P.M.; Brandelli, A. Encapsulation of probiotics and nutraceuticals: Applications in functional food industry. Trends Food Sci. Tech. 2021, 114, 1–10. [Google Scholar] [CrossRef]
- Zeng, W.; Guo, L.; Xu, S.; Chen, J.; Zhou, J. High-throughput screening technology in industrial biotechnology. Trends Biotechnol. 2020, 38, 888–906. [Google Scholar] [CrossRef] [PubMed]
- Tanasupawat, S.; Shida, O.; Okada, S.; Komagata, K. Lactobacillus acidipiscis sp. Nov. and Weissella thailandensis sp. Nov., isolated from fermented fish in Thailand. Int. J. Syst. Evol. Micr. 2000, 50 Pt 4, 1479. [Google Scholar] [CrossRef]
- Aarti, C.; Khusro, A.; Varghese, R.; Arasu, M.V.; Agastian, P.; Al-Dhabi, N.A.; Ilavenil, S.; Choi, K.C. In vitro studies on probiotic and antioxidant properties of Lactobacillus brevis strain LAP2 isolated from Hentak, a fermented fish product of North-East India. LWT 2017, 86, 438–446. [Google Scholar] [CrossRef]
- Liu, J.; Lin, C.; Zhang, W.; Yang, Q.; Meng, J.; He, L.; Deng, L.; Zeng, X. Exploring the bacterial community for starters in traditional high-salt fermented Chinese fish (Suanyu). Food Chem. 2021, 358, 129863. [Google Scholar] [CrossRef] [PubMed]
- Kittibunchakul, S.; Yuthaworawit, N.; Whanmek, K.; Suttisansanee, U.; Santivarangkna, C. Health beneficial properties of a novel plant-based probiotic drink produced by fermentation of brown rice milk with GABA-producing Lactobacillus pentosus isolated from Thai pickled weed. J. Funct. Foods 2021, 86, 104710. [Google Scholar] [CrossRef]
- Luz, C.; Calpe, J.; Manuel Quiles, J.; Torrijos, R.; Vento, M.; Gormaz, M.; Mañes, J.; Meca, G. Probiotic characterization of Lactobacillus strains isolated from breast milk and employment for the elaboration of a fermented milk product. J. Funct. Foods 2021, 84, 104599. [Google Scholar] [CrossRef]
- Hameed, A.M.; Elkhtab, E.; Mostafa, M.S.; Refaey, M.M.M.; Hassan, M.A.A.; Abo El-Naga, M.Y.; Hegazy, A.A.; Rabie, M.M.; Alrefaei, A.F.; Abdulaziz Alfi, A.; et al. Amino acids, solubility, bulk density and water holding capacity of novel freeze-dried cow’s skimmed milk fermented with potential probiotic Lactobacillus plantarum Bu-Eg5 and Lactobacillus rhamnosus Bu-Eg6. Arab J. Chem. 2021, 14, 103291. [Google Scholar] [CrossRef]
- Ong, L.; Shah, N.P. Release and identification of angiotensin-converting enzyme-inhibitory peptides as influenced by ripening temperatures and probiotic adjuncts in Cheddar cheeses. LWT 2008, 41, 1555–1566. [Google Scholar] [CrossRef]
- Tang, W.; Li, C.; He, Z.; Pan, F.; Pan, S.; Wang, Y. Probiotic properties and cellular antioxidant activity of Lactobacillus plantarum MA2 isolated from Tibetan kefir grains. Probiotics Antimicrob. Proteins 2018, 10, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Jeong, D.; Kim, D.; Kang, I.; Kim, H.; Song, K.; Kim, H.; Seo, K. Characterization and antibacterial activity of a novel exopolysaccharide produced by Lactobacillus kefiranofaciens DN1 isolated from kefir. Food Control 2017, 78, 436–442. [Google Scholar] [CrossRef]
- Talib, N.; Mohamad, N.E.; Yeap, S.K.; Hussin, Y.; Aziz, M.N.M.; Masarudin, M.J.; Sharifuddin, S.A.; Hui, Y.W.; Ho, C.L.; Alitheen, N.B. Isolation and characterization of Lactobacillus spp. from kefir samples in Malaysi. Molecules 2019, 24, 2606. [Google Scholar] [CrossRef] [Green Version]
- Fei, Y.; Liu, L.; Liu, D.; Chen, L.; Tan, B.; Fu, L.; Li, L. Investigation on the safety of Lactobacillus amylolyticus L6 and its fermentation properties of tofu whey. LWT 2017, 84, 314–322. [Google Scholar] [CrossRef]
- He, Y. The Isolation and Identification of Lactic Acid Bacteria from Naturally Fermented Tofu Whey and the Application of the Strains in the Acidic Whey Tofu. Master’s Thesis, Jiangnan University, Wuxi, China, 2019. [Google Scholar]
- Aljuobori, A.; Abdullah, N.; Zulkifli, I.; Soleimani, A.F.; Liang, J.B.; Oskoueian, E. Lactobacillus salivarius fermentation reduced glucosinolate and fibre in canola meal. J. Food Res. 2014, 3, 95–102. [Google Scholar] [CrossRef]
- Li, M.; Xu, Y.; Zhang, J. Effect of lactic acid bacteria fermentation on the quality of starchy foods. China Brew. 2020, 39, 13–18. [Google Scholar] [CrossRef]
- Han, M.; Wang, X.; Zhang, M.; Ren, Y.; Yue, T.; Gao, Z. Effect of mixed Lactobacillus on the physicochemical properties of cloudy apple juice with the addition of polyphenols-concentrated solution. Food Biosci. 2021, 41, 101049. [Google Scholar] [CrossRef]
- Yang, X.; Zhou, J.; Fan, L.; Qin, Z.; Chen, Q.; Zhao, L. Antioxidant properties of a vegetable–fruit beverage fermented with two Lactobacillus plantarum strains. Food Sci. Biotechnol. 2018, 27, 1719–1726. [Google Scholar] [CrossRef]
- Jans, C.; Lagler, S.; Lacroix, C.; Meile, L.; Stevens, M.J.A. Complete genome sequences of Lactobacillus curvatus KG6, L. Curvatus MRS6, and Lactobacillus sakei FAM18311, isolated from fermented meat products. Genome Announc. 2017, 5, e00915-17. [Google Scholar] [CrossRef]
- Chaillou, S.P.; Monique, M.C.S.; Dudez, A.; Martin, V.R.; Beaufils, S.; Re, E.D.; Bossy, R.; Loux, V.; Zagorec, M. The complete genome sequence of the meat-borne lactic acid bacterium Lactobacillus sakei 23K. Nat. Biotechnol. 2005, 23, 1527–1533. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Briges, M. The classification of Lactobacilli by means of physiological tests. J. Gen. Microbiol. 1953, 9, 234–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, E.F.; Luciano, W.A.; Xavier, D.E.; Da Costa, W.C.A.; de Sousa Oliveira, K.; Franco, O.L.; de Morais Júnior, M.A.; Lucena, B.T.L.; Picão, R.C.; Magnani, M.; et al. Identification of lactic acid bacteria in fruit pulp processing byproducts and potential probiotic properties of selected Lactobacillus strains. Front. Microbiol. 2016, 7, 1371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riaz Rajoka, M.S.; Mehwish, H.M.; Siddiq, M.; Haobin, Z.; Zhu, J.; Yan, L.; Shao, D.; Xu, X.; Shi, J. Identification, characterization, and probiotic potential of Lactobacillus rhamnosus isolated from human milk. LWT 2017, 84, 271–280. [Google Scholar] [CrossRef]
- Karami, S.; Roayaei, M.; Hamzavi, H.; Bahmani, M.; Hassanzad-Azar, H.; Leila, M.; Rafieian-Kopaei, M. Isolation and identification of probiotic from local dairy and evaluating their antagonistic effect on pathogens. Int. J. Pharm. Investig. 2017, 7, 137–141. [Google Scholar] [CrossRef]
- Anisimova, E.A.; Yarullina, D.R. Antibiotic resistance of Lactobacillus strains. Curr. Microbiol. 2019, 76, 1407–1416. [Google Scholar] [CrossRef]
- Saroj, S.D.; Maudsdotter, L.; Tavares, R.; Jonsson, A.-B. Lactobacilli interfere with Streptococcus pyogenes hemolytic activity and adherence to host epithelial cells. Front. Microbiol. 2016, 7, 1176. [Google Scholar] [CrossRef] [Green Version]
- Alamdary, S.Z.; Bakhshi, B. Lactobacillus acidophilus attenuates toxin production by Vibrio cholerae and shigella dysenteriae following intestinal epithelial cells infection. Microb. Pathog. 2020, 149, 104543. [Google Scholar] [CrossRef]
- Boricha, A.A.; Shekh, S.L.; Pithv, S.P.; Ambalam, P.S.; Vyas, B.R.M. In vitro evaluation of probiotic properties of Lactobacillus species of food and human origin. LWT 2019, 106, 201–208. [Google Scholar] [CrossRef]
- Kieliszek, M.; Pobiega, K.; Piwowarek, K.; Kot, A.M. Characteristics of the proteolytic enzymes produced by lactic acid bacteria. Molecules 2021, 26, 1858. [Google Scholar] [CrossRef]
- Larsen, N.; de Souza, C.B.; Krych, L.; Kot, W.; Leser, T.D.; Sørensen, O.B.; Blennow, A.; Venema, K.; Jespersen, L. Effect of potato fiber on survival of Lactobacillus species at simulated gastric conditions and composition of the gut microbiota in vitro. Food Res. Int. 2019, 125, 108644. [Google Scholar] [CrossRef] [PubMed]
- Larsen, N.; Cahú, T.B.; Isay Saad, S.M.; Blennow, A.; Jespersen, L. The effect of pectins on survival of probiotic Lactobacillus spp. in gastrointestinal juices is related to their structure and physical properties. Food Microbiol. 2018, 74, 11–20. [Google Scholar] [CrossRef]
- Andrade, R.; Santos, E.; Azoubel, P.; Ribeiro, E. Increased survival of Lactobacillus rhamnosus ATCC 7469 in guava juices with simulated gastrointestinal conditions during refrigerated storage. Food Biosci. 2019, 32, 100470. [Google Scholar] [CrossRef]
- Liao, N.; Luo, B.; Gao, J.; Li, X.; Zhao, Z.; Zhang, Y.; Ni, Y.; Tian, F. Oligosaccharides as co-encapsulating agents: Effect on oral Lactobacillus fermentum survival in a simulated gastrointestinal tract. Biotechnol. Lett. 2019, 41, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.H.; Kim, Y.; Kim, J.; Jeong, Y.; Park, H.M.; Kim, J.W.; Kim, J.; Kim, H.; Paek, N.; Kang, C. Evaluating the cryoprotective encapsulation of the lactic acid bacteria in simulated gastrointestinal conditions. Biotechnol. Bioprocess Eng. 2020, 25, 287–292. [Google Scholar] [CrossRef]
- Fijałkowski, K.; Peitler, D.; Rakoczy, R.; Żywicka, A. Survival of probiotic lactic acid bacteria immobilized in different forms of bacterial cellulose in simulated gastric juices and bile salt solution. LWT 2016, 68, 322–328. [Google Scholar] [CrossRef]
- Xu, M.; Zhong, F.; Zhu, J. Evaluating metabolic response to light exposure in Lactobacillus species via targeted metabolic profiling. J. Microbiol. Methods 2017, 133, 14–19. [Google Scholar] [CrossRef]
- Parlindungan, E.; May, B.; Jones, O. Metabolic insights into the effects of nutrient stress on Lactobacillus plantarum B21. Front. Mol. Biosci. 2019, 6, 75. [Google Scholar] [CrossRef] [Green Version]
- Hosseini Nezhad, M.; Hussain, M.A.; Britz, M.L. Stress responses in probiotic Lactobacillus casei. Crit. Rev. Food Sci. 2015, 55, 740–749. [Google Scholar] [CrossRef]
- Palomino, M.M.; Waehner, P.M.; Fina Martin, J.; Ojeda, P.; Malone, L.; Sánchez Rivas, C.; Prado Acosta, M.; Allievi, M.C.; Ruzal, S.M. Influence of osmotic stress on the profile and gene expression of surface layer proteins in Lactobacillus acidophilus ATCC 4356. Appl. Microbiol. Biot. 2016, 100, 8475–8484. [Google Scholar] [CrossRef]
- Samak, G.; Rao, R.; Rao, R. Lactobacillus casei and epidermal growth factor prevent osmotic stress-induced tight junction disruption in caco-2 cell monolayers. Cells 2021, 10, 3578. [Google Scholar] [CrossRef] [PubMed]
- Carasi, P.; Ambrosis, N.; De Antoni, G.; Bressollier, P.; Urdaci, M.; Serradell, M. Adhesion properties of potentially probiotic Lactobacillus kefiri to gastrointestinal mucus. J. Dairy Res. 2014, 81, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Klopper, K.B.; Deane, S.M.; Dicks, L.M.T. Aciduric strains of Lactobacillus reuteri and Lactobacillus rhamnosus, isolated from Human Feces, have strong adhesion and aggregation properties. Probiotics Antimicrob. Proteins 2018, 10, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Sophatha, B.; Piwat, S.; Teanpaisan, R. Adhesion, anti-adhesion and aggregation properties relating to surface charges of selected Lactobacillus strains: Study in Caco-2 and H357 cells. Arch. Microbiol. 2020, 202, 1349–1357. [Google Scholar] [CrossRef]
- Rocha-Mendoza, D.; Kosmerl, E.; Miyagusuku-Cruzado, G.; Giusti, M.M.; Jiménez-Flores, R.; García-Cano, I. Growth of lactic acid bacteria in milk phospholipids enhances their adhesion to Caco-2 cells. J. Dairy Sci. 2020, 103, 7707–7718. [Google Scholar] [CrossRef]
- Zavala, L.; Golowczyc, M.A.; Vandamme, P.; Abraham, A.G. Selected Lactobacillus strains isolated from sugary and milk kefir reduce Salmonella infection of epithelial cells in vitro. Benef. Microbes 2016, 7, 585–595. [Google Scholar] [CrossRef]
- Chen, C.; Lai, C.; Huang, H.; Huang, W.; Toh, H.; Weng, T.; Chuang, Y.; Lu, Y.; Tang, H.-J. Antimicrobial activity of Lactobacillus species against carbapenem-resistant Enterobacteriaceae. Front. Microbiol. 2019, 10, 789. [Google Scholar] [CrossRef]
- de Souza Rodrigues, J.Z.; Passos, M.R.; de Macêdo Neres, N.S.; Almeida, R.S.; Pita, L.S.; Santos, I.A.; Silveira, P.H.S.; Reis, M.M.; Santos, I.P.; de Oliveira Negrão Ricardo, L.; et al. Antimicrobial activity of Lactobacillus fermentum TcUESC01 against Streptococcus mutans UA159. Microb. Pathog. 2020, 142, 104063. [Google Scholar] [CrossRef]
- Dubourg, G.; Elsawi, Z.; Raoult, D. Assessment of the in vitro antimicrobial activity of Lactobacillus species for identifying new potential antibiotics. Int. J. Antimicrob. Agents 2015, 46, 590–593. [Google Scholar] [CrossRef]
- Sornsenee, P.; Singkhamanan, K.; Sangkhathat, S.; Saengsuwan, P.; Romyasamit, C. Probiotic properties of Lactobacillus species isolated from fermented palm sap in Thailand. Probiotics Antimicrob. Proteins 2021, 13, 957–969. [Google Scholar] [CrossRef]
- Aguirre, L.; Hebert, E.M.; Garro, M.S.; de Giori, G.S. Proteolytic activity of Lactobacillus strains on soybean proteins. LWT 2014, 59, 780–785. [Google Scholar] [CrossRef]
- Esteban-Torres, M.; Mancheño, J.M.; de Las Rivas, B.; Muñoz, R. Characterization of a halotolerant lipase from the lactic acid bacteria Lactobacillus plantarum useful in food fermentations. LWT 2015, 60, 246–252. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Xie, Y.; Yu, Z.; Meng, G.; Wu, Z. Effect of Lactobacillus plantarum expressing multifunctional glycoside hydrolases on the characteristics of alfalfa silage. Appl. Microbiol. Biotechnol. 2019, 103, 7983–7995. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Yu, H.; Feng, X.; Tang, H.; Xiong, Z.; Xia, Y.; Ai, L.; Song, X. Specific bile salt hydrolase genes in Lactobacillus plantarum AR113 and relationship with bile salt resistance. LWT 2021, 145, 111208. [Google Scholar] [CrossRef]
- Rai, A.K.; Sanjukta, S.; Jeyaram, K. Production of angiotensin I converting enzyme inhibitory (ACE-I) peptides during milk fermentation and their role in reducing hypertension. Crit. Rev. Food Sci. 2017, 57, 2789–2800. [Google Scholar] [CrossRef]
- Moslehishad, M.; Ehsani, M.R.; Salami, M.; Mirdamadi, S.; Ezzatpanah, H.; Naslaji, A.N.; Moosavi-Movahedi, A.A. The comparative assessment of ACE-inhibitory and antioxidant activities of peptide fractions obtained from fermented camel and bovine milk by Lactobacillus rhamnosus PTCC 1637. Int. Dairy J. 2013, 29, 82–87. [Google Scholar] [CrossRef]
- Vanessa Moraes Ramalho Castro, M.D.M.S.; Guerra, A.F.; Riger, C.J.; Laureano-Melo, R.; Luchese, R.H. Role of milk and honey in the tolerance of Lactobacilli to oxidative stress. Braz. J. Microbiol. 2021, 52, 883–893. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, C.; Li, D.; Zhao, Y.; Zhang, X.; Zeng, X.; Yang, Z.; Li, S. Antioxidant activity of an exopolysaccharide isolated from Lactobacillus plantarum C88. Int. J. Biol. Macromol. 2013, 54, 270–275. [Google Scholar] [CrossRef]
- Ahmadi, M.A.; Ebrahimi, M.T.; Mehrabian, S.; Tafvizi, F.; Bahrami, H.; Dameshghian, M. Antimutagenic and anticancer effects of lactic acid bacteria isolated from Tarhana through Ames test and phylogenetic analysis by 16S rDNA. Nutr. Cancer 2014, 66, 1406–1413. [Google Scholar] [CrossRef]
- Pepoyan, A.Z.; Balayan, M.H.; Malkhasyan, L.; Manvelyan, A.; Bezhanyan, T.; Paronikyan, R.; Tsaturyan, V.V.; Tatikyan, S.; Kamiya, S.; Chikindas, M.L. Effects of probiotic Lactobacillus acidophilus strain INMIA 9602 Er 317/402 and putative probiotic Lactobacilli on DNA damages in the small intestine of Wistar Rats in vivo. Probiotics Antimicrob. Proteins 2019, 11, 905–909. [Google Scholar] [CrossRef]
- Tukenmez, U.; Aktas, B.; Aslim, B.; Yavuz, S. The relationship between the structural characteristics of Lactobacilli-EPS and its ability to induce apoptosis in colon cancer cells in vitro. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amiri, S.; Rezaei Mokarram, R.; Sowti Khiabani, M.; Rezazadeh Bari, M.; Alizadeh Khaledabad, M. In situ production of conjugated linoleic acid by Bifidobacterium lactis BB12 and Lactobacillus acidophilus LA5 in milk model medium. LWT 2020, 132, 109933. [Google Scholar] [CrossRef]
- Ameen, F.A.; Hamdan, A.M.; El-Naggar, M.Y. Assessment of the heavy metal bioremediation efficiency of the novel marine lactic acid bacterium, Lactobacillus plantarum MF042018. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Son, S.; Jeon, H.; Jeon, E.B.; Lee, N.; Park, Y.; Kang, D.; Paik, H. Potential probiotic Lactobacillus plantarum Ln4 from kimchi: Evaluation of β-galactosidase and antioxidant activities. LWT 2017, 85, 181–186. [Google Scholar] [CrossRef]
- Chamberlain, C.A.; Hatch, M.; Garrett, T.J. Metabolomic profiling of oxalate-degrading probiotic Lactobacillus acidophilus and Lactobacillus gasseri. PLoS ONE 2019, 14, e0222393. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Gu, Q.; Yang, L.; Yu, Y.; Wang, Y. Characterization of extracellular vitamin B12 producing Lactobacillus plantarum strains and assessment of the probiotic potentials. Food Chem. 2017, 234, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Markowiak-Kopeć, P.; Śliżewska, K. The effect of probiotics on the production of short-chain fatty acids by human intestinal microbiome. Nutrients 2020, 12, 1107. [Google Scholar] [CrossRef] [Green Version]
- Hamzehlou, P.; Sepahy, A.A.; Mehrabian, S.; Hosseini, F. Production of vitamins B3, B6 and B9 by Lactobacillus Isolated from traditional yogurt samples from 3 cities in Iran, Winter 2016. Appl. Food Biotechnol. 2018, 5, 107–120. [Google Scholar] [CrossRef]
- Bourdichon, F.; Laulund, S.; Tenning, P. Inventory of microbial species with a rationale: A comparison of the IDF/EFFCA inventory of microbial food cultures with the EFSA Biohazard Panel qualified presumption of safety. FEMS Microbiol. Lett. 2019, 366, fnz048. [Google Scholar] [CrossRef]
- Tsai, C.; Lai, C.; Yu, B.; Tsen, H. Use of PCR primers and probes based on the 23S rRNA and internal transcription spacer (its) gene sequence for the detection and enumerization of Lactobacillus acidophilus and Lactobacillus plantarum in feed supplements. Anaerobe 2010, 16, 270–277. [Google Scholar] [CrossRef]
- Singh, H.; Kongo, J.M.; Borges, A.; Ponte DJ, B.; Griffiths, M.W. Lactic acid bacteria isolated from raw milk cheeses: Ribotyping, antimicrobial activity against selected food pathogens and resistance to common antibiotics. J. Food Process. Technol. 2015, 6, 485. [Google Scholar] [CrossRef]
- Galanis, A.; Kourkoutas, Y.; Tassou, C.C.; Chorianopoulos, N. Detection and identification of probiotic Lactobacillus plantarum strains by multiplex PCR using RAPD-derived primers. Int. J. Mol. Sci. 2015, 16, 25141–25153. [Google Scholar] [CrossRef] [PubMed]
- Khemariya, P.; Singh, S.; Jaiswal, N.; Chaurasia, S.N.S. Isolation and ddentification of Lactobacillus plantarum from vegetable samples. Food Biotechnol. 2016, 30, 49–62. [Google Scholar] [CrossRef]
- Kim, E.; Yang, S.; Lim, B.; Park, S.H.; Rackerby, B.; Kim, H. Design of PCR assays to specifically detect and identify 37 Lactobacillus species in a single 96 well plate. BMC Microbiol. 2020, 20, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Jarocki, P.; Komoń-Janczara, E.; Glibowska, A.; Dworniczak, M.; Pytka, M.; Korzeniowska-Kowal, A.; Wzorek, A.; Kordowska-Wiater, M. Molecular routes to specific identification of the Lactobacillus casei group at the species, subspecies and strain level. Int. J. Mol. Sci. 2020, 21, 2694. [Google Scholar] [CrossRef]
- Felis, G.E.; Dellaglio, F.; Mizzi, L.; Torriani, S. Comparative sequence analysis of a recA gene fragment brings new evidence for a change in the taxonomy of the Lactobacillus casei group. Int. J. Syst. Evol. Micr. 2001, 51, 2113–2117. [Google Scholar] [CrossRef] [Green Version]
- Rodas, A.M.; Ferrer, S.; Pardo, I.Y. Polyphasic study of wine Lactobacillus strains: Taxonomic implications. Int. J. Syst. Evol. Microbiol. 2005, 55, 197–207. [Google Scholar] [CrossRef]
- Adesulu-Dahunsi, A.T.; Sanni, A.I.; Jeyaram, K.; Banwo, K. Genetic diversity of Lactobacillus plantarum strains from some indigenous fermented foods in Nigeria. LWT 2017, 82, 199–206. [Google Scholar] [CrossRef]
- Lu, W.; Kong, W.; Yang, P.; Kong, J. A one-step PCR-based method for specific identification of 10 common lactic acid bacteria and Bifidobacterium in fermented milk. Int. Dairy J. 2015, 41, 7–12. [Google Scholar] [CrossRef]
- Maleki Kakelar, H.; Barzegari, A.; Hanifian, S.; Barar, J.; Omidi, Y. Isolation and molecular identification of Lactobacillus with probiotic potential from abomasums driven rennet. Food Chem. 2019, 272, 709–714. [Google Scholar] [CrossRef]
- Kaur, J.; Sharma, A.; Lee, S.; Park, Y. Molecular typing of Lactobacillus brevis isolates from Korean food using repetitive element-polymerase chain reaction. Food Sci. Technol. Int. 2018, 24, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Liu, T.; Zhang, G.; Chen, J.; Gu, J.; Yuan, L.; He, G. Genotyping of Lactobacillus sanfranciscensis isolates from Chinese traditional sourdoughs by multilocus sequence typing and multiplex RAPD-PCR. Int. J. Food Microbiol. 2017, 258, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, C.; Bancalari, E.; Milanović, V.; Cardinali, F.; Osimani, A.; Sardaro, M.L.S.; Bottari, B.; Bernini, V.; Aquilanti, L.; Clementi, F.; et al. Study of the bacterial diversity of foods: PCR-DGGE versus LH-PCR. Int. J. Food Microbiol. 2017, 242, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Pasulka, A.L.; Howes, A.L.; Kallet, J.G.; VanderKelen, J.; Villars, C. Visualization of probiotics via epifluorescence microscopy and fluorescence in situ hybridization (FISH). J. Microbiol. Methods 2021, 182, 106151. [Google Scholar] [CrossRef]
- Chatzopoulou, S.; Eriksson, N.L.; Eriksson, D. Improving risk assessment in the European Food Safety Authority: Lessons from the European Medicines Agency. Front. Plant Sci. 2020, 11, 349. [Google Scholar] [CrossRef] [Green Version]
- Morris, C.P.; Bergman, Y.; Tekle, T.; Fissel, J.A.; Tamma, P.D.; Simner, P.J.; Burnham, C.D. Cefiderocol antimicrobial susceptibility testing against multidrug-resistant Gram-negative bacilli: A comparison of disk diffusion to broth microdilution. J. Clin. Microbiol. 2020, 59, e01649-20. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, E.; van Westreenen, M.; Goessens, W.; Croughs, P. The accuracy of four commercial broth microdilution tests in the determination of the minimum inhibitory concentration of colistin. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 1–8. [Google Scholar] [CrossRef]
- Kumar, S.; Pattanaik, A.K.; Jadhav, S.E. Potent health-promoting effects of a synbiotic formulation prepared from Lactobacillus acidophilus NCDC15 fermented milk and Cichorium intybus root powder in Labrador dogs. Curr. Res. Biotechnol. 2021, 3, 209–214. [Google Scholar] [CrossRef]
- Chiu, S.; Chen, C.; Wang, L.; Huang, L. Whole-genome sequencing of Lactobacillus salivarius strains BCRC 14759 and BCRC 12574. Genome Announc. 2017, 5, e01336-17. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, J.; Zhang, D.; Liu, H.; Wang, S.; Wang, Y.; Ji, H. Complete genome sequencing and comparative genome characterization of Lactobacillus johnsonii ZLJ010, a potential probiotic with health-promoting properties. Front. Genet. 2019, 10, 812. [Google Scholar] [CrossRef] [Green Version]
- Pasolli, E.; De Filippis, F.; Mauriello, I.E.; Cumbo, F.; Walsh, A.M.; Leech, J.; Cotter, P.D.; Segata, N.; Ercolini, D. Large-scale genome-wide analysis links lactic acid bacteria from food with the gut microbiome. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Sánchez, S.; Fernández-Pacheco, P.; Seseña, S.; Pintado, C.; Palop, M.L. Selection of probiotic Lactobacillus strains with antimicrobial activity to be used as biocontrol agents in food industry. LWT 2021, 143, 111142. [Google Scholar] [CrossRef]
- Geng, T.; He, F.; Su, S.; Sun, K.; Zhao, L.; Zhao, Y.; Bao, N.; Pan, L.; Sun, H. Probiotics Lactobacillus rhamnosus GG ATCC53103 and Lactobacillus plantarum JL01 induce cytokine alterations by the production of TCDA, DHA, and succinic and palmitic acids, and enhance immunity of weaned piglets. Res. Vet. Sci. 2021, 137, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Zhu, X.; Tuo, Y.; Zhang, H.; Li, Y.; Xu, C.; Mu, G.; Jiang, S. Reducing antigenicity of β-lactoglobulin, probiotic properties and safety evaluation of Lactobacillus plantarum AHQ-14 and Lactobacillus bulgaricus BD0390. Food Biosci. 2021, 42, 101137. [Google Scholar] [CrossRef]
- Melini, F.; Melini, V.; Luziatelli, F.; Ficca, A.G.; Ruzzi, M. Health-promoting components in fermented foods: An up-to-date systematic review. Nutrients 2019, 11, 1189. [Google Scholar] [CrossRef] [Green Version]
- de Oliveira, P.M.; Santos, L.P.; Coelho, L.F.; Avila Neto, P.M.; Sass, D.C.; Contiero, J. Production of L (+) Lactic Acid by Lactobacillus casei Ke11: Fed batch fermentation strategies. Fermentation 2021, 7, 151. [Google Scholar] [CrossRef]
- Allgeyer, L.C.; Miller, M.J.; Lee, S.Y. Sensory and microbiological quality of yogurt drinks with prebiotics and probiotics. J. Dairy Sci. 2010, 93, 4471–4479. [Google Scholar] [CrossRef]
- Tang, S.; Cheng, Y.; Wu, T.; Hu, F.; Pan, S.; Xu, X. Effect of Lactobacillus plantarum-fermented mulberry pomace on antioxidant properties and fecal microbial community. LWT 2021, 147, 111651. [Google Scholar] [CrossRef]
- Lv, X.; Chen, M.; Huang, Z.; Guo, W.; Ai, L.; Bai, W.; Yu, X.; Liu, Y.; Rao, P.; Ni, L. Potential mechanisms underlying the ameliorative effect of Lactobacillus paracasei FZU103 on the lipid metabolism in hyperlipidemic mice fed a high-fat diet. Food Res. Int. 2021, 139, 109956. [Google Scholar] [CrossRef]
- Stearns, J.C.; Lynch, M.D.J.; Senadheera, D.B.; Tenenbaum, H.C.; Goldberg, M.B.; Cvitkovitch, D.G.; Croitoru, K.; Moreno-Hagelsieb, G.; Neufeld, J.D. Bacterial biogeography of the human digestive tract. Sci. Rep. 2011, 1, 170. [Google Scholar] [CrossRef] [Green Version]
- Xiao, L.; Feng, Q.; Liang, S.; Sonne, S.B.; Xia, Z.; Qiu, X.; Li, X.; Long, H.; Zhang, J.; Zhang, D.; et al. A catalog of the mouse gut metagenome. Nat. Biotechnol. 2015, 33, 1103–1108. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.-N.C.; Wu, H.A.; DE Srjhfb Chen, Y.-Z.; Chen, Y.-J.; Shen, X.-Z.; Liu, T.-T. Altered molecular signature of intestinal microbiota in irritable bowel syndrome patients compared with healthy controls: A systematic review and meta-analysis. Digest. Liver Dis. 2017, 49, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Poles, M.; Fisch, G.; Ma, Y.; Nossa, C.; Phelan, J.; Pei, Z. HIV induced immunosuppression is associated with colonization of the proximal gut by environmental bacteria. AIDS 2016, 30, 19–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karlsson, F.H.; Tremaroli, V.; Nookaew, I.; Bergström, G.; Behre, C.J.; Fagerberg, B.; Nielsen, J.; Bäckhed, F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 2013, 498, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Forslund, K.; Hildebrand, F.; Nielsen, T.; Falony, G.; Le Chatelier, E.; Sunagawa, S.; Prifti, E.; Vieira-Silva, S.; Gudmundsdottir, V.; Krogh Pedersen, H. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 2015, 528, 262–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khalesi, S.; Sun, J.; Buys, N.; Jayasinghe, R. Effect of probiotics on blood pressure. Hypertension 2014, 64, 897–903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmadi, E.; Alizadeh-Navaei, R.; Rezai, M.S. Efficacy of probiotic use in acute rotavirus diarrhea in children: A systematic review and meta-analysis. Caspian J. Intern. Med. 2015, 6, 187–195. [Google Scholar]
- Peng, Y.; Li, A.; Yu, L.; Qin, G. The role of probiotics in prevention and treatment for patients with allergic rhinitis: A systematic review. Am. J. Rhinol. Allergy 2015, 29, 292–298. [Google Scholar] [CrossRef]
- Szajewska, H.; Ruszczyński, M.; Radzikowski, A. Probiotics in the prevention of antibiotic-associated diarrhea in children: A meta-analysis of randomized controlled trials. J. Pediatr. 2006, 149, 367–372.e1. [Google Scholar] [CrossRef]
- Zhang, M.; Qian, W.; Qin, Y.; He, J.; Zhou, Y. Probiotics in helicobacter pylori eradication therapy: A systematic review and meta-analysis. World J. Gastroenterol. 2015, 21, 4345–4357. [Google Scholar] [CrossRef]
- Jang, Y.J.; Kim, W.; Han, D.H.; Lee, K.; Ko, G. Lactobacillus fermentum species ameliorate dextran sulfate sodium-induced colitis by regulating the immune response and altering gut microbiota. Gut Microbes 2019, 10, 696–711. [Google Scholar] [CrossRef]
- Li, X.; Wang, N.; Yin, B.; Fang, D.; Zhao, J.; Zhang, H.; Wang, G.; Chen, W. Lactobacillus plantarum X1 with α-glucosidase inhibitory activity ameliorates type 2 diabetes in mice. Rsc. Adv. 2016, 6, 63536–63547. [Google Scholar] [CrossRef]
- Lim, E. Evaluation of the anti-Helicobacter pylori and cytotoxic properties of the antimicrobial substances from Lactobacillus acidophilus BK13 and Lactobacillus paracasei BK57. Korean J. Microbiol. 2015, 51, 156–168. [Google Scholar] [CrossRef] [Green Version]
- Rajoka, M.R.R.; Zhao, H.; Lu, Y.; Lian, Z.; Li, N.; Hussain, N.; Shao, D.; Jin, M.; Li, Q.; Shi, J. Anticancerpotential against cervix cancer (HeLa) cell line of probiotic Lactobacillus casei and Lactobacillus paracasei strains isolated from human breast milk. Food Funct. 2018, 9, 2705–2715. [Google Scholar] [CrossRef] [PubMed]
- Suo, H.; Liu, S.; Li, J.; Ding, Y.; Wang, H.; Zhang, Y.; Zhao, X.; Song, J. Lactobacillus paracasei ssp. Paracasei YBJ01 reduced d-galactose-induced oxidation in male Kuming mice. J. Dairy Sci. 2018, 101, 10664–10674. [Google Scholar] [CrossRef] [Green Version]
- Lee, I.; Caggianiello, G.; van Swam, I.I.; Taverne, N.; Meijerink, M.; Bron, P.A.; Spano, G.; Kleerebezem, M. Strain-specific features of extracellular polysaccharides and their impact on Lactobacillus plantarum-host interactions. Appl. Environ. Microbiol. 2016, 82, 3959–3970. [Google Scholar] [CrossRef] [Green Version]
- Sanhueza, E.; Paredes-Osses, E.; González, C.L.; García, A. Effect of pH in the survival of Lactobacillus salivarius strain UCO_979C wild type and the pH acid acclimated variant. Electron. J. Biotechnol. 2015, 18, 343–346. [Google Scholar] [CrossRef] [Green Version]
- Zhu, M.; Xie, R.; Chen, L.; You, M.; Gou, W.; Chen, C.; Li, P.; Cai, Y. Milk production and quality of lactating yak fed oat silage prepared with a low-temperature-tolerant lactic acid bacteria inoculant. Foods 2021, 10, 2437. [Google Scholar] [CrossRef]
- Zotta, T.; Parente, E.; Ricciardi, A. Aerobic metabolism in the genus Lactobacillus: Impact on stress response and potential applications in the food industry. J. Appl. Microbiol. 2017, 122, 857–869. [Google Scholar] [CrossRef] [Green Version]
- Vázquez, J.A.; Mirón, J.; González, M.P.; Murado, M.A. Effects of aeration on growth and on production of bacteriocins and other metabolites in cultures of eight strains of lactic acid bacteria. Appl. Biochem. Biotechnol. 2005, 127, 111–124. [Google Scholar] [CrossRef]
- Tian, X.; Wang, Y.; Chu, J.; Mohsin, A.; Zhuang, Y. Exploring cellular fatty acid composition and intracellular metabolites of osmotic-tolerant mutant Lactobacillus paracasei NCBIO-M2 for highly efficient lactic acid production with high initial glucose concentration. J. Biotechnol. 2018, 286, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Panwar, D.; Kapoor, M. Transcriptional analysis of galactomannooligosaccharides utilization by Lactobacillus plantarum WCFS1. Food Microbiol. 2020, 86, 103336. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Hwang, C.E.; Cho, E.J.; Song, Y.H.; Kim, S.C.; Cho, K.M. Improvement of nutritional components and in vitro antioxidative properties of soy-powder yogurts using Lactobacillus plantarum. J. Food Drug Anal. 2018, 26, 1054–1065. [Google Scholar] [CrossRef] [PubMed]
- Glušac, J.; Stijepić, M.; Đurđević-Milošević, D.; Milanović, S.; Kanurić, K.; Vukić, V. Growth and viability of Lactobacillus delbrueckii subsp. bulgaricus and Streptococcus thermophilus in traditional yoghurt enriched by honey and whey protein concentrate. Iran J. Vet. Res. 2015, 16, 249–254. [Google Scholar] [PubMed]
- Guimarães, A.; Santiago, A.; Teixeira, J.A.; Venâncio, A.; Abrunhosa, L. Anti-aflatoxigenic effect of organic acids produced by Lactobacillus plantarum. Int. J. Food Microbiol. 2018, 264, 31–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villegas, J.M.; Brown, L.; Savoy De Giori, G.; Hebert, E.M. Optimization of batch culture conditions for GABA production by Lactobacillus brevis CRL 1942, isolated from quinoa sourdough. LWT 2016, 67, 22–26. [Google Scholar] [CrossRef]
- Jiang, Y.; Yang, Z. A functional and genetic overview of exopolysaccharides produced by Lactobacillus plantarum. J. Funct. Foods 2018, 47, 229–240. [Google Scholar] [CrossRef]
- Bachmann, H.; Pronk, J.T.; Kleerebezem, M.; Teusink, B. Evolutionary engineering to enhance starter culture performance in food fermentations. Curr. Opin. Biotech. 2015, 32, 1–7. [Google Scholar] [CrossRef]
- Ito, M.; Kim, Y.; Tsuji, H.; Takahashi, T.; Kiwaki, M.; Nomoto, K.; Danbara, H.; Okada, N. Transposon mutagenesis of probiotic Lactobacillus casei identifies asnH, an asparagine synthetase gene involved in its immune-activating capacity. PLoS ONE 2014, 9, e83876. [Google Scholar] [CrossRef] [Green Version]
- Perpetuini, G.; Scornec, H.; Tofalo, R.; Serror, P.; Schirone, M.; Suzzi, G.; Corsetti, A.; Cavin, J.F.; Licandro-Seraut, H. Identification of critical genes for growth in olive brine by transposon mutagenesis of Lactobacillus pentosus C11. Appl. Environ. Microbiol. 2013, 79, 4568–4575. [Google Scholar] [CrossRef]
- Joshi, D.S.; Singhvi, M.S.; Khire, J.M.; Gokhale, D.V. Strain improvement of Lactobacillus lactis for D-lactic acid production. Biotechnol. Lett. 2010, 32, 517–520. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Chen, X.; Yu, L.; Xu, W.; Wang, P.; Zhang, X.; Li, W.; Li, C.; Ren, N. Screening of Lactobacillus rhamnosus strains mutated by microwave irradiation for increased lactic acid production. Afr. J. Microbiol. Res. 2012, 6, 6055–6065. [Google Scholar] [CrossRef]
- Iang, A.L.; Hu, W.; Li, W.J.; Liu, L.; Tian, X.J.; Liu, J.; Wang, S.Y.; Lu, D.; Chen, J.H. Enhanced production of L-lactic acid by Lactobacillus thermophiles SRZ50 mutant generated by high-linear energy transfer heavy ion mutagenesis. Eng. Life Sci. 2018, 18, 626–634. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Li, Q.; Wang, S.; Bai, J.; Dong, M.; Xiao, G.; Wang, J. Lactobacillus casei JY300-8 generated by 12C6+ beams mutagenesis inhibits tumor progression by modulating the gut microbiota in mice. J. Funct. Foods 2021, 87, 104779. [Google Scholar] [CrossRef]
- Gao, C.; Yang, F.; Liu, Y. Plasma mutation breeding of high yield γ-aminobutyric acid lactic acid bacteria. Gene Sci. Eng. 2017, 1, 8–18. [Google Scholar] [CrossRef]
- Chen, Y.; Tian, X.W.; Li, Q.H.; Li, Y.; Zhuang, Y.P. Target-site directed rational high-throughput screening system for high sophorolipids production by Candida bombicola. Bioresour. Technol. 2020, 315, 123856–123864. [Google Scholar] [CrossRef]
- Lv, X.; Song, J.; Yu, B.; Liu, H.; Li, C.; Zhuang, Y.; Wang, Y. High-throughput system for screening of high L-lactic acid productivity strains in deep-well microtiter plates. Bioprocess Biosyst. Eng. 2016, 39, 1737–1747. [Google Scholar] [CrossRef]
- Hugenholtz, J.; Sybesma, W.; Groot, M.N.; Wisselink, W.; Ladero, V.; Burgess, K.; van Sinderen, D.; Piard, J.; Eggink, G.; Smid, E.J.; et al. Metabolic engineering of lactic acid bacteria for the production of nutraceuticals. In Lactic Acid Bacteria: Genetics, Metabolism and Applications, 2nd ed.; Siezen, R.J., Kok, J., Abee, T., Schaafsma, G., Eds.; Springer: Berlin, Germany, 2002; Volume 82, pp. 217–235. [Google Scholar]
- Pan, H.; Zhan, J.; Yang, H.; Wang, C.; Liu, H.; Zhou, H.; Zhou, H.; Lu, X.; Su, X.; Tian, Y. Improving the acid resistance of Tannase TanBLp (AB379685) from Lactobacillus plantarum ATCC14917T by site-specific mutagenesis. Indian J. Microbiol. 2022, 62, 96–102. [Google Scholar] [CrossRef]
- Tian, X.; Liu, X.; Zhang, Y.; Chen, Y.; Hang, H.; Chu, J.; Zhuang, Y. Metabolic engineering coupled with adaptive evolution strategies for the efficient production of high-quality L-lactic acid by Lactobacillus paracasei. Bioresour. Technol. 2021, 323, 124549. [Google Scholar] [CrossRef]
- Upadhyaya, B.P.; DeVeaux, L.C.; Christopher, L.P. Metabolic engineering as a tool for enhanced lactic acid production. Trends Biotechnol. 2014, 32, 637–644. [Google Scholar] [CrossRef]
- Tian, X.; Chen, H.; Liu, H.; Chen, J. Recent advances in lactic acid production by lactic acid bacteria. Appl. Biochem. Biotech. 2021, 193, 4151–4171. [Google Scholar] [CrossRef] [PubMed]
- Papagianni, M. Metabolic engineering of lactic acid bacteria for the production of industrially important compounds. Comput. Struct. Biotec. 2012, 3, e201210003. [Google Scholar] [CrossRef] [Green Version]
- Ye, L.; Zhao, H.; Li, Z.; Wu, J.C. Improved acid tolerance of Lactobacillus pentosus by error-prone whole genome amplification. Bioresour. Technol. 2013, 135, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Hospet, R.; Thangadurai, D.; Cruz-Martins, N.; Sangeetha, J.; Appaiah, K.A.A.; Chowdhury, Z.Z.; Bedi, N.; Soytong, K.; Al Tawahaj, A.R.M.; Jabeen, S.; et al. Genome shuffling for phenotypic improvement of industrial strains through recursive protoplast fusion technology. Crit. Rev. Food Sci. 2021, 1, 1–10. [Google Scholar] [CrossRef]
- Sun, J.; Liu, H.; Dang, L.; Liu, J.; Wang, J.; Lu, Z.; Lu, Y. Genome shuffling of Lactobacillus plantarum 163 enhanced antibacterial activity and usefulness in preserving orange juice. Lett. Appl. Microbiol. 2021, 73, 741–749. [Google Scholar] [CrossRef] [PubMed]
- van Pijkeren, J.P.; Britton, R.A. Precision genome engineering in lactic acid bacteria. Microb. Cell Factories 2014, 13, S10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Börner, R.A.; Kandasamy, V.; Axelsen, A.M.; Nielsen, A.T.; Bosma, E.F. Genome editing of lactic acid bacteria: Opportunitie for food, feed, pharma and biotech. FEMS Microbiol. Lett. 2019, 366, fny291. [Google Scholar] [CrossRef]
- Plavec, T.V.; Berlec, A. Safety aspects of genetically modified lactic acid bacteria. Microorganisms 2020, 8, 297. [Google Scholar] [CrossRef] [Green Version]
- Panesar, P.S. Fermented dairy products: Starter cultures and potential nutritional benefits. Food Nutr. Sci. 2011, 2, 47–51. [Google Scholar] [CrossRef] [Green Version]
- Goveas, L.C.; Ashwath, K.S.; Nazerath, B.R.; Dsouza, O.; Ullekh; Umesh, A.; Muddappa, V.S. Development of coconut water-based exopolysaccharide rich functional beverage by fermentation with probiotic Lactobacillus plantarum SVP2. Biocatal. Agric. Biotechnol. 2021, 34, 102030. [Google Scholar] [CrossRef]
- Szutowska, J. Functional properties of lactic acid bacteria in fermented fruit and vegetable juices: A systematic literature review. Eur. Food Res. Technol. 2020, 246, 357–372. [Google Scholar] [CrossRef]
- Crispín-Isidro, G.; Lobato-Calleros, C.; Espinosa-Andrews, H.; Alvarez-Ramirez, J.; Vernon-Carter, E.J. Effect of inulin and agave fructans addition on the rheological, microstructural and sensory properties of reduced-fat stirred yogurt. LWT 2016, 62, 438–444. [Google Scholar] [CrossRef]
- Ozogul, F.; Ozcelik, S.; Ozogul, Y.; Yilmaz, M.T. Seafood infusion broths as novel sources to produce organic acids using selected lactic acid bacteria strains. Food Biosci 2021, 43, 101227. [Google Scholar] [CrossRef]
- Siedler, S.; Balti, R.; Neves, A.R. Bioprotective mechanisms of lactic acid bacteria against fungal spoilage of food. Curr. Opin. Biotech. 2019, 56, 138–146. [Google Scholar] [CrossRef]
- Papizadeh, M.; Rohani, M.; Nahrevanian, H.; Hosseini, S.N.; Shojaosadati, S.A.; Pourshafie, M.R. Using various approaches of design of experiments for high cell density production of the functionally probiotic Lactobacillus plantarum strain RPR42 in a cane molasses-based medium. Curr. Microbiol. 2020, 77, 1756–1766. [Google Scholar] [CrossRef] [PubMed]
- E, J.; Ma, L.; Chen, Z.; Ma, R.; Zhang, Q.; Sun, R.; He, Z.; Wang, J. Effects of buffer salts on the freeze-drying survival rate of Lactobacillus plantarum LIP-1 based on transcriptome and proteome analyses. Food Chem. 2020, 326, 126849. [Google Scholar] [CrossRef]
- Singhvi, M.; Zendo, T.; Sonomoto, K. Free lactic acid production under acidic conditions by lactic acid bacteria strains: Challenges and future prospects. Appl. Microbiol. Biot. 2018, 102, 5911–5924. [Google Scholar] [CrossRef]
- Bähr, C.; Leuchtle, B.; Lehmann, C.; Becker, J.; Jeude, M.; Peinemann, F.; Arbter, R.; Büchs, J. Dialysis shake flask for effective screening in fed-batch mode. Biochem. Eng. J. 2012, 69, 182–195. [Google Scholar] [CrossRef]
- Costas Malvido, M.; Alonso González, E.; Pérez Guerra, N. Nisin production in realkalized fed-batch cultures in whey with feeding with lactose- or glucose-containing substrates. Appl. Microbiol. Biotechnol. 2016, 100, 7899–7908. [Google Scholar] [CrossRef]
- Kawai, M.; Tsuchiya, A.; Ishida, J.; Yoda, N.; Yashiki-Yamasaki, S.; Katakura, Y. Suppression of lactate production in fed-batch culture of some lactic acid bacteria with sucrose as the carbon source. J. Biosci. Bioeng. 2020, 129, 535–540. [Google Scholar] [CrossRef]
- Hu, J.; Lin, Y.; Zhang, Z.; Xiang, T.; Mei, Y.; Zhao, S.; Liang, Y.; Peng, N. High-titer lactic acid production by Lactobacillus pentosus FL0421 from corn stover using fed-batch simultaneous saccharification and fermentation. Bioresour. Technol. 2016, 214, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Sadiq, F.A.; Xu, Z.; Liu, Z.; Zhao, J.; Zhang, H.; Chen, W. High-density cultivation of Lactobacillus and Bifidobacterium using an automatic feedback feeding method. LWT 2019, 112, 108232. [Google Scholar] [CrossRef]
- Zhao, L.; Tang, Z.; Gu, Y.; Shan, Y.; Tang, T. Investigate the cross-flow flat-plate photobioreactor for high-density culture of microalgae. Asia-Pac. J. Chem. Eng. 2018, 13, e2247. [Google Scholar] [CrossRef]
- Chen, P.; Hong, Z.; Cheng, C.; Ng, I.; Lo, Y.; Nagarajan, D.; Chang, J. Exploring fermentation strategies for enhanced lactic acid production with polyvinyl alcohol-immobilized Lactobacillus plantarum 23 using microalgae as feedstock. Bioresour. Technol. 2020, 308, 123266. [Google Scholar] [CrossRef]
- Cha, K.H.; Lee, E.H.; Yoon, H.S.; Lee, J.H.; Kim, J.Y.; Kang, K.; Park, J.; Jin, J.B.; Ko, G.; Pan, C. Effects of fermented milk treatment on microbial population and metabolomic outcomes in a three-stage semi-continuous culture system. Food Chem. 2018, 263, 216–224. [Google Scholar] [CrossRef]
- Gao, M.J.; Nie, Y.F.; Zhang, H.X.; Xie, Y.H.; Hao, H.W.; Liu, H. Application of directed vat set yogurt starters by space Lactobacillus reuteri. Nat. Prod. Res. Dev. 2019, 31, 136–142. [Google Scholar] [CrossRef]
- Fonseca, F.; Cenard, S.; Passot, S. Freeze-drying of lactic acid bacteria. In Cryopreservation and Freeze-Drying Protocols, 1st ed.; Wolkers, W., Oldenhof, H., Eds.; Springer: New York, NY, USA, 2015; Volume 1257, pp. 477–488. [Google Scholar] [CrossRef]
- Gallardo-Rivera, C.; Báez-González, J.G.; García-Alanís, K.G.; Torres-Alvarez, C.; Dares-Sánchez, K.; Szymanski, A.; Amaya-Guerra, C.A.; Castillo, S. Effect of three types of drying on the viability of lactic acid bacteria in Foam-Mat dried yogurt. Processes 2021, 9, 2123. [Google Scholar] [CrossRef]
- Moreira, M.T.C.; Martins, E.; Perrone, Í.T.; de Freitas, R.; Queiroz, L.S.; de Carvalho, A.F. Challenges associated with spray drying of lactic acid bacteria: Understanding cell viability loss. Compr. Rev. Food Sci. Food Saf. 2021, 20, 3267–3283. [Google Scholar] [CrossRef]
- Hao, F.; Fu, N.; Ndiaye, H.; Woo, M.W.; Jeantet, R.; Chen, X.D. Thermotolerance, survival, and stability of lactic acid bacteria after spray drying as affected by the increase of growth temperature. Food Bioprocess Tech. 2021, 14, 120–132. [Google Scholar] [CrossRef]
- Lu, W.; Fu, N.; Woo, M.W.; Chen, X.D. Exploring the interactions between Lactobacillus rhamnosus GG and whey protein isolate for preservation of the viability of bacteria through spray drying. Food Funct. 2021, 12, 2995–3008. [Google Scholar] [CrossRef]
- Mamun, A.A.; Payap, M.; Jaruwan, M. Viability of Lactobacillus plantarum TISTR 2083 in protectant during low-temperature drying and storage. Sains Malays. 2021, 50, 2227–2238. [Google Scholar] [CrossRef]
- Shanmugam, S. Granulation techniques and technologies: Recent progresses. Bioimpacts 2015, 5, 55–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vishali, D.A.; Monisha, J.; Sivakamasundari, S.K.; Moses, J.A.; Anandharamakrishnan, C. Spray freeze drying: Emerging applications in drug delivery. J. Control. Release 2019, 300, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Choi, I.S.; Ko, S.H.; Kim, H.M.; Chun, H.H.; Lee, K.H.; Yang, J.E.; Jeong, S.; Park, H.W. Shelf-life extension of freeze-dried Lactobacillus brevis WiKim0069 using supercooling pretreatment. LWT 2019, 112, 108230. [Google Scholar] [CrossRef]




| Fermented Products | Lactobacilli Species Present | References |
|---|---|---|
| Fermented fish products | L. acidipiscis, L. brevis, L. delbrueckii, L. fermentum, L. pentosus, L. plantarum, L. versmoldensis | [25,26,27] |
| Fermented dairy products | L. delbrueckii, L. bulgaricus, L. fermentum, L. plantarum, L. kefiri, L. paracasei subsp. Paracasei, L. rhamnosus, L. curvatus | [6,28,29,30,31,32,33,34] |
| Fermented soy products | L. amylophilus, L. buchneri, L. delbrueckii, L. fermentum, L. paracasei, L. plantarum, L. salivarius | [35,36,37] |
| Fermented starch foods | L. acidophilus, L. brevis, L. casei, L. bulgaricus, L. oryzae, L. pentosus, L. reuteri, L. rhamnosus, L. rossiae, L. sakei, L. curvatus, L. panis, L. sanfranciscensis | [38] |
| Fermented fruit and vegetable | L. acidophilus, L. brevis, L. casei, L. fermentum, L. pentosus, L. plantarum | [28,39,40] |
| Fermented meat products | L. sakei, L. curvatus | [41,42] |
| Characteristic | Assays | Representative References |
|---|---|---|
| Safety | Strain identification (including physiological and biochemical tests, molecular level) | [44,45,46,47] |
| Antibiotic resistance | [48] | |
| Hemolytic activity | [49] | |
| Determination of potential metabolites (enzyme production, toxin production, production of biogenic amines) | [50,51,52] | |
| Tolerance to stress | Low pH and bile (for example, artificial gastric and pancreatic juices and GIT simulators) | [17,53,54,55,56,57,58] |
| Growth environment (for example, nutrition substrate, osmotic pressure, light, temperature, oxygen) | [59,60,61,62] | |
| Adhesion ability | Cell surface hydrophobicity | [63] |
| Adhesion to mucus (for example, adhesion to mucin) | [64,65,66] | |
| Adhesion to Caco-2/TC7 cells | [67,68] | |
| Antimicrobial activity | Production of antimicrobial metabolites such as lactic acid and bacteriocin against pathogenic bacteria (e.g., streak methods, disk diffusion methods, turbidimetric assays, biofluorescence analysis) | [69,70,71] |
| Autoaggregation, Coaggregation | [18,72] | |
| Technological properties | Proteolytic activity (e.g., production of various proteases) | [73] |
| Lipolytic activity (e.g., production of lipases) | [74] | |
| Carbohydrate degradation activity (e.g., production of various glycosidases, amylases, cellulases) | [75] | |
| Reduce cardiovascular disease | Cholesterol degradation tests (e.g., Bile salt hydrolase activity) | [76] |
| Metabolites such as peptides inhibit the ACE activity | [77,78] | |
| Antioxidant | Tolerance to hydrogen peroxide | [79] |
| Metabolites such as the antioxidant activity of extracellular polysaccharides, peptides | [78,80] | |
| Anticancer | Ames test | [81] |
| Comet assay | [82] | |
| Nitrosamine degradation assay | [9] | |
| Inducing apoptosis of cancer cells test | [83] | |
| Additional characteristics | Conjugated linoleic acid test | [84] |
| The removal of heavy metals | [85] | |
| β-Galactosidase activity analysis | [86] | |
| Determination of oxalic acid degradation | [87] | |
| Determination of production of short-chain unsaturated fatty acids and vitamins | [88,89,90] |
| Methods Used | Comments | Species Identified and Source | Reference |
|---|---|---|---|
| 23S rDNA probe | Probes unequivocally differentiated L. acidophilus and L. plantarum isolates. | L. acidophilus, L. pentosus, L. plantarum species isolates from feed supplements or starter products | [92] |
| Ribotyping | Good discrimination at strains level based upon differences in rRNA. | Some L. paracasei ss. paracasei strains as the dominant ones from raw milk cheeses | [93] |
| RAPD | Good discrimination at strains level. | L. plantarum 2035 and L. plantarum ACA-DC 2640 isolated from Feta cheese | [94] |
| Species-specific PCR (plantaricin biosynthesis protein gene) | Rapid and preliminary screening of L. plantarum from large vegetable samples before performing a battery of phenotypic and molecular methods. | L. plantarum from vegetable samples | [95] |
| Species-specific PCR using 16S rRNA or unique genes primers | Successful in the species detected in 17 products matched those indicated on their labels, whereas the remaining products contained species other than those appearing on the label. | Some Lactobacillus spp., 19 probiotics and 12 dairy products | [96] |
| Genus- and species-specific PCR, multiplex PCR, real-time HRM analysis, RFLP-PCR, rep-PCR, RAPD-PCR, AFLP-PCR, and proteomic methods such as MALDI-TOF MS typing and SDS-PAGE fingerprinting | Multiplex PCR and MALDI-TOF MS were the most valuable methods to identify the tested bacteria at the species level. At the strain level, the AFLP-PCR method showed the highest discriminatory power. | L. casei group, two international collections of microorganisms—the Japan Collection of Microorganisms (JCM) and Belgian Coordinated Collections of Microorganisms (BCCM) | [97] |
| Comparative sequence analysis, stretches of rec A gene | Successful in a clear separation of all type strains in distinct branches; identification of L. casei ATCC 393 and L. casei ATCC 334 as L. zeae and L. paracasei, respectively. | L. casei, L. paracasei (both subspecies), L. rhamnosus, L. zeae, strains from a commercial probiotic product. | [98] |
| 16S ARDRA, RAPD, Eco RI ribotyping | 13 wine strains typed as L. paracasei/casei, based on similar band pattern as L. paracasei type strain and L. casei ATCC 334. | L. casei/L. paracasei from wine | [99] |
| PFGE | Good discrimination at strain level based upon different bacterial strains. | The strains of L. plantarum isolated from the different fermented foods | [100] |
| One-step PCR-based, using 16S rRNA genes primers | Successful differentiation among 10 common lactic acid bacteria at the species level. | L. delbrueckii and others from fermented milk | [101] |
| 16S ARDRA, ribotyping, RAPD | Only RAPD and ribotyping could discriminate between the type strains of both species. | L. plantarum, L. pentosus, Wine isolates | [99] |
| PCR-ARDRA (Taq I), RAPD | ARDRA and RAPD approaches may demonstrate a robust efficiency in the discrimination of unknown isolates. | L. acidophilus, L. planetarum, and L. fermentum from abomasums driven rennet | [102] |
| Repetitive-element PCR | Could rapidly and easily differentiate L. brevis species at strains level. | The closely related strains of L. brevis species | [103] |
| Multi-locus sequence typing (MLST) and multiplex RAPD-PCR | Targeting different genetic variations under the combination of MLST and multiplex-RAPD analysis | L. sanfranciscensis, Chinese traditional sourdoughs | [104] |
| PCR-DGGE, length-heterogeneity PCR (LH-PCR) | Good discrimination at strains level. | Type and reference strains of L. brevis DSMZ 20556 and L. plantarum DSMZ 2601 | [105] |
| FISH | Rapid and accurate way to identify and quantify bacterial species. | L. plantarum (Probiotic products) | [106] |
| Functional Properties | Example | Reference |
|---|---|---|
| Regulating immune system | Jang et al. evaluated immunometabolic functions of L. fermentum strains (KBL374 and KBL375) isolated from the feces of healthy Koreans. | [133] |
| Regulating the balance of blood glucose, blood lipid, and blood pressure | Li et al. found that L. plantarum X1 can alleviate the symptoms of diabetes by improving the level of short-chain fatty acids in type 2 diabetic mice. | [134] |
| Antimicrobial activity | Lim et al. found that L. paracasei BK 57 has antagonistic effect on Helicobacter pylori and can be used as potential antibiotics. | [135] |
| Lower blood pressure | Ong et al. found that L. paracasei can isolate and purify ACE inhibitory peptides from cheddar cheese. | [31] |
| Antitumor | Rajoka et al. found that the antiproliferative activity of the fermentation supernatant of L. paracasei SR 4 on cervical cancer cells was up to 89%. L. paracasei showed high anti-cancer activity by promoting the up-regulation of BAX, BAD, caspase3, caspase8, and caspase9 genes and down-regulating the expression of the Bcl-2 gene. | [136] |
| Antioxidant | Suo et al. found that L. paracasei ybJ 01 can significantly improve D-galactose, induced the ability of serum superoxide dismutase (SOD), glutathione peroxidase and total antioxidant in mice, and inhibited the production of malondialdehyde. | [137] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Y.; Xing, S.; He, L.; Li, C.; Wang, X.; Zeng, X.; Dai, Y. Characterization, High-Density Fermentation, and the Production of a Directed Vat Set Starter of Lactobacilli Used in the Food Industry: A Review. Foods 2022, 11, 3063. https://doi.org/10.3390/foods11193063
Lu Y, Xing S, He L, Li C, Wang X, Zeng X, Dai Y. Characterization, High-Density Fermentation, and the Production of a Directed Vat Set Starter of Lactobacilli Used in the Food Industry: A Review. Foods. 2022; 11(19):3063. https://doi.org/10.3390/foods11193063
Chicago/Turabian StyleLu, Yun, Shuqi Xing, Laping He, Cuiqin Li, Xiao Wang, Xuefeng Zeng, and Yifeng Dai. 2022. "Characterization, High-Density Fermentation, and the Production of a Directed Vat Set Starter of Lactobacilli Used in the Food Industry: A Review" Foods 11, no. 19: 3063. https://doi.org/10.3390/foods11193063
APA StyleLu, Y., Xing, S., He, L., Li, C., Wang, X., Zeng, X., & Dai, Y. (2022). Characterization, High-Density Fermentation, and the Production of a Directed Vat Set Starter of Lactobacilli Used in the Food Industry: A Review. Foods, 11(19), 3063. https://doi.org/10.3390/foods11193063