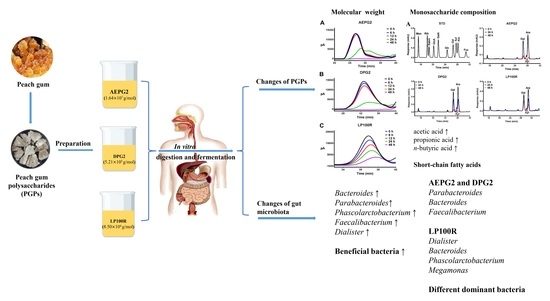
In Vitro Digestion and Fecal Fermentation of Peach Gum Polysaccharides with Different Molecular Weights and Their Impacts on Gut Microbiota
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation and Physicochemical Properties of PGPs
2.3. Simulated In Vitro Digestion of PGPs
2.4. In Vitro Simulated Fermentation of PGPs
2.4.1. Preparation of Human Fecal Microbiota
2.4.2. PGPs Fermentation In Vitro
2.4.3. Determination of Chemical Indices in the Fermentation Broth during Fermentation
2.4.4. Determination of SCFA Content during Fermentation
2.4.5. Determination of the Composition of Gut Microbiota
2.5. Statistical Analysis
3. Results and Discussion
3.1. Monosaccharide Composition and Molecular Weight of PGPs
3.2. Characteristics of PGPs during In Vitro Digestion
3.3. Characteristics of PGPs during In Vitro Fermentation
3.3.1. Carbohydrate Consumption during Fermentation
3.3.2. Changes in the pH of the Fermentation Broth
3.3.3. Changes in the Molecular Weights of PGPs during Fermentation
3.3.4. Changes in Monosaccharide Composition of PGPs during Fermentation
3.3.5. Changes in SCFA Contents
3.4. Effect of PGPs on Microbial Communities
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Simas-Tosin, F.F.; Wagner, R.; Santos, E.M.R.; Sassaki, G.L.; Gorin, P.A.J.; Iacomini, M. Polysaccharide of nectarine gum exudate: Comparison with that of peach gum. Carbohyd. Polym. 2009, 76, 485–487. [Google Scholar] [CrossRef]
- Simas, F.F.; Gorin, P.A.J.; Wagner, R.; Sassaki, G.L.; Bonkerner, A.; Iacomini, M. Comparison of structure of gum exudate polysaccharides from the trunk and fruit of the peach tree (Prunus persica). Carbohyd. Polym. 2008, 71, 218–228. [Google Scholar] [CrossRef]
- Wei, C.Y.; He, P.F.; He, L.; Ye, X.Q.; Cheng, J.W.; Wang, Y.B.; Li, W.Q.; Liu, Y. Structure characterization and biological activities of a pectic polysaccharide from cupule of Castanea henryi. Int. J. Biol. Macromol. 2018, 109, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.H.; Tan, J.S.; Wang, G.; Zhou, L. Superior amine-rich gel adsorbent from peach gum polysaccharide for highly efficient removal of anionic dyes. Carbohyd. Polym. 2018, 199, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.H.; Tan, J.S.; Xu, X.; Huang, X.H.; Zhou, L. Facile synthesis of amphiphilic peach gum polysaccharide as a robust host for efficient encapsulation of methylene blue and methyl orange dyes from water. Int. J. Biol. Macromol. 2020, 154, 974–980. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.H.; Long, J.W.; Sun, J.H.; Wang, G.; Zhou, L. A review on peach gum polysaccharide: Hydrolysis, structure, properties and applications. Carbohyd. Polym. 2022, 279, 119015. [Google Scholar] [CrossRef]
- Wu, D.T.; Fu, Y.; Guo, H.; Yuan, Q.; Nie, X.R.; Wang, S.P.; Gan, R.Y. In vitro simulated digestion and fecal fermentation of polysaccharides from loquat leaves: Dynamic changes in physicochemical properties and impacts on human gut microbiota. Int. J. Biol. Macromol. 2021, 168, 733–742. [Google Scholar] [CrossRef]
- Wu, D.T.; An, L.Y.; Liu, W.; Hu, Y.C.; Wang, S.P.; Zhou, L. In vitro fecal fermentation properties of polysaccharides from Tremella fuciformis and related modulation effects on gut microbiota. Food Res. Int. 2022, 156, 111185. [Google Scholar] [CrossRef]
- Cummings, J.H. Short chain fatty acids in the human colon. Gut 1981, 22, 763–779. [Google Scholar] [CrossRef] [Green Version]
- Lovegrove, A.; Edwards, C.H.; De Noni, I.; Patel, H.; El, S.N.; Grassby, T.; Zielke, C.; Ulmius, M.; Nilsson, L.; Butterworth, P.J.; et al. Role of polysaccharides in food, digestion, and health. Crit. Rev. Food Sci. 2017, 57, 237–253. [Google Scholar] [CrossRef]
- Song, Q.Q.; Wang, Y.K.; Huang, L.X.; Shen, M.Y.; Yu, Y.; Yu, Q.; Chen, Y.; Xie, J.H. Review of the relationships among polysaccharides, gut microbiota, and human health. Food Res. Int. 2021, 140, 109858. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.Y.; Zhang, Y.; He, L.; Cheng, J.W.; Li, J.H.; Tao, W.Y.; Mao, G.Z.; Zhang, H.; Linhardt, R.J.; Ye, X.Q.; et al. Structural characterization and anti-proliferative activities of partially degraded polysaccharides from peach gum. Carbohyd. Polym. 2019, 203, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.Y.; Zhang, Y.; Zhang, H.; Li, J.H.; Tao, W.Y.; Linhardt, R.J.; Chen, S.G.; Ye, X.Q. Physicochemical properties and conformations of water-soluble peach gums via different preparation methods. Food Hydrocoll. 2019, 95, 571–579. [Google Scholar] [CrossRef]
- Minekus, M.; Alminger, M.; Alvito, P.; Ballance, S.; Bohn, T.; Bourlieu, C.; Carriere, F.; Boutrou, R.; Corredig, M.; Dupont, D.; et al. A standardised static in vitro digestion method suitable for food—an international consensus. Food Funct. 2014, 5, 1113–1124. [Google Scholar] [CrossRef] [Green Version]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Li, W.; Wang, K.Q.; Sun, Y.; Ye, H.; Hu, B.; Zeng, X.X. Influences of structures of galactooligosaccharides and fructooligosaccharides on the fermentation in vitro by human intestinal microbiota. J. Funct. Foods 2015, 13, 158–168. [Google Scholar] [CrossRef]
- Zhou, W.T.; Yan, Y.M.; Mi, J.; Zhang, H.C.; Lu, L.; Luo, Q.; Li, X.Y.; Zeng, X.X.; Cao, Y.L. Simulated digestion and fermentation in vitro by human gut microbiota of polysaccharides from Bee collected pollen of Chinese Wolfberry. J. Agr. Food Chem. 2018, 66, 898–907. [Google Scholar] [CrossRef]
- Dubois, M.; Gilles, K.; Hamilton, J.K.; Rebers, P.A.; Smith, F. A colorimetric method for the determination of sugars. Nature 1951, 168, 167. [Google Scholar] [CrossRef]
- Ding, Q.A.; Nie, S.P.; Hu, J.L.; Zong, X.Y.; Li, Q.Q.; Xie, M.Y. In vitro and in vivo gastrointestinal digestion and fermentation of the polysaccharide from Ganoderma atrum. Food Hydrocoll. 2017, 63, 646–655. [Google Scholar] [CrossRef]
- Li, X.J.; Guo, R.; Wu, X.J.; Liu, X.; Ai, L.Z.; Sheng, Y.; Song, Z.B.; Wu, Y. Dynamic digestion of tamarind seed polysaccharide: Indigestibility in gastrointestinal simulations and gut microbiota changes in vitro. Carbohyd. Polym. 2020, 239, 116194. [Google Scholar] [CrossRef]
- Li, S.J.; Li, M.X.; Yue, H.; Zhou, L.S.; Huang, L.L.; Du, Z.Y.; Ding, K. Structural elucidation of a pectic polysaccharide from Fructus mori and its bioactivity on intestinal bacteria strains. Carbohyd. Polym. 2018, 186, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.T.; He, Y.; Yuan, Q.; Wang, S.P.; Gan, R.Y.; Hu, Y.C.; Zou, L. Effects of molecular weight and degree of branching on microbial fermentation characteristics of okra pectic-polysaccharide and its selective impact on gut microbial composition. Food Hydrocoll. 2022, 132, 107897. [Google Scholar] [CrossRef]
- Ding, Y.; Yan, Y.M.; Peng, Y.J.; Chen, D.; Mi, J.; Lu, L.; Luo, Q.; Li, X.Y.; Zeng, X.X.; Cao, Y.L. In vitro digestion under simulated saliva, gastric and small intestinal conditions and fermentation by human gut microbiota of polysaccharides from the fruits of Lycium barbarum. Int. J. Biol. Macromol. 2019, 125, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.L.; Nie, S.P.; Min, F.F.; Xie, M.Y. Artificial simulated saliva, gastric and intestinal digestion of polysaccharide from the seeds of Plantago asiatica L. Carbohyd. Polym. 2013, 92, 1143–1150. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The role of short-chain fatty acids in health and disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar] [PubMed]
- Patnode, M.L.; Beller, Z.W.; Han, N.D.; Cheng, J.Y.; Peters, S.L.; Terrapon, N.; Henrissat, B.; Le Gall, S.; Saulnier, L.; Hayashi, D.K.; et al. Interspecies competition impacts targeted manipulation of human gut bacteria by fiber-derived glycans. Cell 2019, 179, 59–73. [Google Scholar] [CrossRef]
- Hou, Z.Q.; Hu, X.X.; Luan, L.Q.; Yu, C.X.; Wang, X.X.; Chen, S.G.; Ye, X.Q. Prebiotic potential of RG-I pectic polysaccharides from Citrus subcompressa by novel extraction methods. Food Hydrocoll. 2022, 124. [Google Scholar] [CrossRef]
- Wang, K.; Liao, M.F.; Zhou, N.; Bao, L.; Ma, K.; Zheng, Z.Y.; Wang, Y.J.; Liu, C.; Wang, W.Z.; Wang, J.; et al. Parabacteroides distasonis alleviates obesity and metabolic dysfunctions via production of succinate and secondary bile acids. Cell Rep. 2019, 26, 222–235. [Google Scholar] [CrossRef] [Green Version]
- Szentirmai, E.; Millican, N.S.; Massie, A.R.; Kapas, L. Butyrate, a metabolite of intestinal bacteria, enhances sleep. Sci. Rep. 2019, 9, 7035. [Google Scholar] [CrossRef] [Green Version]
- Zou, X.; Ji, J.; Qu, H.; Wang, J.; Shu, D.M.; Wang, Y.; Liu, T.F.; Li, Y.; Luo, C.L. Effects of sodium butyrate on intestinal health and gut microbiota composition during intestinal inflammation progression in broilers. Poult. Sci. 2019, 98, 4449–4456. [Google Scholar] [CrossRef]
- Sakamoto, M.; Ikeyama, N.; Toyoda, A.; Murakami, T.; Mori, H.; Iino, T.; Ohkuma, M. Dialister hominis sp. nov., isolated from human faeces. Int. J. Syst. Evol. Micr. 2020, 70, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Al-Lahham, S.H.; Peppelenbosch, M.P.; Roelofsen, H.; Vonk, R.J.; Venema, K. Biological effects of propionic acid in humans; metabolism, potential applications and underlying mechanisms. Biochim. Biophys. Acta 2010, 1801, 1175–1183. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.Q.; Zhu, Y.; Li, X.T.; Sun, B.G. Dynamic balancing of intestinal short-chain fatty acids: The crucial role of bacterial metabolism. Trends Food Sci. Tech. 2020, 100, 118–130. [Google Scholar] [CrossRef]
- Vacca, M.; Celano, G.; Calabrese, F.M.; Portincasa, P.; Gobbetti, M.; De Angelis, M. The controversial role of human gut Lachnospiraceae. Microorganisms 2020, 8, 573. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, D.K.; Renuka; Dangi, A.K.; Shandilya, U.K.; Puniya, A.K.; Shukla, P. Chapter 44 –New-generation probiotics: Perspectives and applications. In Microbiome and Metabolome in Diagnosis, Therapy, and other Strategic Applications; Faintuch, J., Faintuch, S., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 417–424. [Google Scholar]
- Liu, C.Y.; Sun, Y.Y.; Jia, Y.Q.; Geng, X.Q.; Pan, L.C.; Jiang, W.; Xie, B.Y.; Zhu, Z.Y. Effect of steam explosion pretreatment on the structure and bioactivity of Ampelopsis grossedentata polysaccharides. Int. J. Biol. Macromol. 2021, 185, 194–205. [Google Scholar] [CrossRef] [PubMed]
- Hashizume, K.; Tsukahara, T.; Yamada, K.; Koyama, H.; Ushida, K. Megasphaera elsdenii JCM1772T normalizes hyperlactate production in the large intestine of fructooligosaccharide-fed rats by stimulating butyrate production. J. Nutr. 2003, 133, 3187–3190. [Google Scholar] [CrossRef]







| Sample | Monosaccharide Composition (mol %) | Mw (g/mol) | ||||||
|---|---|---|---|---|---|---|---|---|
| Mannose | Rhamnose | Glucuronic Acid | Glucose | Galactose | Xylose | Arabinose | ||
| AEPG2 | 3.08 ± 0.06 c | 0.73 ± 0.03 b | 3.08 ± 0.06 a | 1.54 ± 0.10 a | 34.95 ± 0.80 c | 7.20 ± 0.10 a | 49.41 ± 1.50 a | (1.64 ± 0.04) 107 a |
| DPG2 | 4.64 ± 0.09 b | 1.02 ± 0.06 a | 2.61 ± 0.09 b | trace | 39.82 ± 0.51 a | 3.89 ± 0.06 b | 48.02 ± 1.4 b | (5.21 ± 0.41) 105 b |
| LP100R | 5.42 ± 0.11 a | 1.00 ± 0.05 a | 2.70 ± 0.09 b | trace | 35.80 ± 0.43 b | 7.04 ± 0.25 a | 48.21 ± 1.25 b | (8.5 ± 0.6) 104 c |
| Groups | Indices | |||||
|---|---|---|---|---|---|---|
| Total OTUs | Total Tags | Goods Coverage | Chao1 | Shannon | Simpson | |
| Blank | 1105 ± 24.02 a | 74,878 ± 2335 e | 1.00 ± 0.00 a | 1192.94 ± 57.68 ab | 5.73 ± 0.03 a | 0.95 ± 0.01 ab |
| GOS | 400 ± 65.83 e | 77,712 ± 2411 d | 1.00 ± 0.00 a | 598.28 ±36.81 d | 3.84 ± 0.13 b | 0.87 ± 0.01 b |
| AEPG2 | 913 ± 75.48 b | 91,668 ± 4132 b | 1.00 ± 0.00 a | 1295.39 ± 94.02 a | 5.91 ± 0.26 a | 0.96 ± 0.01 a |
| DPG2 | 801 ± 14.64 d | 81,671 ± 2006 c | 1.00 ± 0.00 a | 1101.75 ± 99.58b c | 5.58 ± 0.26 a | 0.95 ± 0.01 a |
| LP100R | 853 ± 87.61 c | 120,167 ± 41,739 a | 1.00 ± 0.00 a | 1028.69 ± 54.38 c | 5.59 ± 0.12 a | 0.96 ± 0.01 a |
| Phylum | Family | Genus | Relative Abundances (%) | ||||
|---|---|---|---|---|---|---|---|
| Blank | GOS | AEPG2 | DPG2 | LP100R | |||
| Actinobacteria | Bifidobacteriaceae | Bifidobacterium | - | 18.94 ± 0.94 a | 0.01 ± 0.00 b | - | - |
| Bacteroidetes | Bacteroidaceae | Bacteroides | 7.19 ± 0.28 e | 10.70 ± 0.40 d | 37.82 ± 0.89 b | 45.83 ± 1.82 a | 34.33 ± 0.64 c |
| Bacteroidetes | Porphyromonadaceae | Parabacteroides | 0.67 ± 0.02 d | 1.31 ± 0.04 c | 2.71 ± 0.16 a | 1.21 ± 0.05 cd | 1.64 ± 0.07 b |
| Firmicutes | Lachnospiraceae | Blautia | 4.35 ± 0.15 a | 0.26 ± 0.01 d | 2.48 ± 0.12 b | 2.13 ± 0.10 c | 2.55 ± 0.12 b |
| Firmicutes | Lachnospiraceae | 9.23 ± 0.42 a | 0.67 ± 0.02 e | 8.34 ± 0.40 b | 6.15 ± 0.29 c | 4.74 ± 0.23 d | |
| Firmicutes | Ruminococcaceae | Faecalibacterium | 5.96 ± 0.21 c | 0.26 ± 0.01 e | 7.93 ± 0.36 a | 7.49 ± 0.31 b | 2.03 ± 0.11 d |
| Firmicutes | Veillonellaceae | Dialister | 1.12 ± 0.05 c | 1.48 ± 0.06 b | 0.47 ± 0.01 e | 0.83 ± 0.02 d | 3.20 ± 0.11 a |
| Firmicutes | Veillonellaceae | Megamonas | 13.04 ± 0.67 b | 19.31 ± 0.93 a | 7.48 ± 0.36 e | 8.62 ± 0.40 d | 12.00 ± 0.49 c |
| Firmicutes | Veillonellaceae | Phascolarctobacterium | 1.03 ± 0.03 c | 0.97 ± 0.03 c | 4.71 ± 0.18 bc | 5.28 ± 0.16 b | 7.58 ± 0.35 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, C.; Yao, L.; Zhang, L.; Zhang, Y.; Luo, Q.; Qiu, S.; Zeng, X.; Chen, S.; Ye, X. In Vitro Digestion and Fecal Fermentation of Peach Gum Polysaccharides with Different Molecular Weights and Their Impacts on Gut Microbiota. Foods 2022, 11, 3970. https://doi.org/10.3390/foods11243970
Wei C, Yao L, Zhang L, Zhang Y, Luo Q, Qiu S, Zeng X, Chen S, Ye X. In Vitro Digestion and Fecal Fermentation of Peach Gum Polysaccharides with Different Molecular Weights and Their Impacts on Gut Microbiota. Foods. 2022; 11(24):3970. https://doi.org/10.3390/foods11243970
Chicago/Turabian StyleWei, Chaoyang, Li Yao, Lin Zhang, Yu Zhang, Qian Luo, Shuyi Qiu, Xiangyong Zeng, Shiguo Chen, and Xingqian Ye. 2022. "In Vitro Digestion and Fecal Fermentation of Peach Gum Polysaccharides with Different Molecular Weights and Their Impacts on Gut Microbiota" Foods 11, no. 24: 3970. https://doi.org/10.3390/foods11243970
APA StyleWei, C., Yao, L., Zhang, L., Zhang, Y., Luo, Q., Qiu, S., Zeng, X., Chen, S., & Ye, X. (2022). In Vitro Digestion and Fecal Fermentation of Peach Gum Polysaccharides with Different Molecular Weights and Their Impacts on Gut Microbiota. Foods, 11(24), 3970. https://doi.org/10.3390/foods11243970