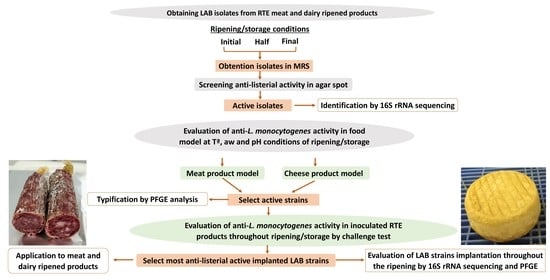
Strategies for Biocontrol of Listeria monocytogenes Using Lactic Acid Bacteria and Their Metabolites in Ready-to-Eat Meat- and Dairy-Ripened Products
Abstract
:1. Introduction
2. Selection and Evaluation of LAB from RTE Meat and Dairy-Ripened Products with Anti-L. monocytogenes Activity
3. Effect of Selected LAB Strains on L. monocytogenes Inhibition
3.1. Production of Inhibitory Compounds
3.2. Competition for Nutrients
3.3. Competition for Space
3.4. Reduction of L. monocytogenes Virulence by LAB
4. Bacteriocins with Activity against L. monocytogenes
Selection of Bacteriocin-Producing Lactic Acid Bacteria and Bacteriocin Characterization
5. Application of Selected LAB or Bacteriocins in RTE Dry-Cured Meat Products
6. Application of Selected LAB or Bacteriocins in RTE Dairy-Ripened Products
7. Conclusions and Future Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baka, M.; Noriega, E.; Mertens, L.; Van Derlinden, E.; Van Impe, J.F.M. Protective role of indigenous Leuconostoc carnosum against Listeria monocytogenes on vacuum packed Frankfurter sausages at suboptimal temperatures. Food Res. Int. 2014, 66, 197–206. [Google Scholar] [CrossRef]
- EFSA; ECDC. The European Union One Health 2019 Zoonoses Report. EFSA J. 2021, 19, 6406. [Google Scholar] [CrossRef]
- EFSA; ECDC. Scientific opinion on the Listeria monocytogenes contamination of ready-to-eat foods and the risk for human health in the EU. EFSA J. 2018, 16, 1–173. [Google Scholar] [CrossRef]
- Kurpas, M.; Wieczorek, K.; Osek, J. Ready-to-eat meat products as a source of Listeria monocytogenes. J. Vet. Res. 2018, 62, 49–55. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Rios, V.; Dalgaard, P. Prevalence of Listeria monocytogenes in European cheeses: A systematic review and meta-analysis. Food Control 2018, 84, 205–214. [Google Scholar] [CrossRef] [Green Version]
- Fretz, R.; Sagel, U.; Ruppitsch, W.; Pietzka, A.T.; Stöger, A.; Huhulescu, S.; Heuberger, S.; Pichler, J.; Much, P.; Pfaff, G.; et al. Listeriosis outbreak caused by acid curd cheese “Quargel”, Austria and Germany 2009. Eurosurveillance 2010, 15, 1–2. [Google Scholar] [CrossRef]
- Cartwright, E.J.; Jackson, K.A.; Johnson, S.D.; Graves, L.M.; Silk, B.J.; Mahon, B.E. Listeriosis outbreaks and associated food vehicles, United States, 1998–2008. Emerg. Infect. Dis. 2013, 19, 1–9. [Google Scholar] [CrossRef]
- Magalhães, R.; Almeida, G.; Ferreira, V.; Santos, I.; Silva, J.; Mendes, M.M.; Pita, J.; Mariano, G.; Mâncio, I.; Sousa, M.M. Cheese-related listeriosis outbreak, Portugal, march 2009 to february 2012. Eurosurveillance 2015, 20, 21104. [Google Scholar] [CrossRef]
- Egan, K.; Field, D.; Rea, M.C.; Ross, R.P.; Hill, C.; Cotter, P.D. Bacteriocins: Novel solutions to age old spore-related problems? Front. Microbiol. 2016, 7, 461. [Google Scholar] [CrossRef] [Green Version]
- Morandi, S.; Silvetti, T.; Battelli, G.; Brasca, M. Can lactic acid bacteria be an efficient tool for controlling Listeria monocytogenes contamination on cheese surface? The case of Gorgonzola cheese. Food Control 2019, 96, 499–507. [Google Scholar] [CrossRef]
- EFSA BIOHAZ Panel. Updated list of QPS status recommended biological agents in support of EFSA risk assessments. EFSA J. 2021, 19, 1–5. [Google Scholar]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Cocolin, L.; Foschino, R.; Comi, G.; Grazia Fortina, M. Description of the bacteriocins produced by two strains of Enterococcus faecium isolated from Italian goat milk. Food Microbiol. 2007, 24, 752–758. [Google Scholar] [CrossRef]
- Sip, A.; Wieckowicz, M.; Olejnik-Schmidt, A.; Grajek, W. Anti-Listeria activity of lactic acid bacteria isolated from golka, a regional cheese produced in Poland. Food Control 2012, 26, 117–124. [Google Scholar] [CrossRef]
- Xiraphi, N.; Georgalaki, M.; Rantsiou, K.; Cocolin, L.; Tsakalidou, E.; Drosinos, E.H. Purification and characterization of a bacteriocin produced by Leuconostoc mesenteroides E131. Meat Sci. 2008, 80, 194–203. [Google Scholar] [CrossRef]
- Campagnollo, F.B.; Margalho, L.P.; Kamimura, B.A.; Feliciano, M.D.; Freire, L.; Lopes, L.S.; Alvarenga, V.O.; Cadavez, V.A.P.; Gonzales-Barron, U.; Schaffner, D.W.; et al. Selection of indigenous lactic acid bacteria presenting anti-listerial activity, and their role in reducing the maturation period and assuring the safety of traditional Brazilian cheeses. Food Microbiol. 2018, 73, 288–297. [Google Scholar] [CrossRef]
- Panebianco, F.; Giarratana, F.; Caridi, A.; Sidari, R.; De Bruno, A.; Giuffrida, A. Lactic acid bacteria isolated from traditional Italian dairy products: Activity against Listeria monocytogenes and modelling of microbial competition in soft cheese. LWT 2021, 137, 110446. [Google Scholar] [CrossRef]
- Papamanoli, E.; Tzanetakis, N.; Litopoulou-Tzanetaki, E.; Kotzekidou, P. Characterization of lactic acid bacteria isolated from a Greek dry-fermented sausage in respect of their technological and probiotic properties. Meat Sci. 2003, 65, 859–867. [Google Scholar] [CrossRef]
- Todorov, S.D.; Dicks, L.M.T. Characterization of bacteriocins produced by lactic acid bacteria isolated from spoiled black olives. J. Basic Microbiol. 2005, 45, 312–322. [Google Scholar] [CrossRef]
- Picozzi, C.; Bonacina, G.; Vigentini, I.; Foschino, R. Genetic diversity in Italian Lactobacillus sanfranciscensis strains assessed by multilocus sequence typing and pulsed-field gel electrophoresis analyses. Microbiology 2010, 156, 2035–2045. [Google Scholar] [CrossRef] [Green Version]
- Park, S.H.; Jung, J.H.; Seo, D.H.; Lee, H.L.; Kim, G.W.; Park, S.Y.; Shin, W.C.; Hong, S.; Park, C.S. Differentiation of lactic acid bacteria based on RFLP analysis of the tuf gene. Food Sci. Biotechnol. 2012, 21, 911–915. [Google Scholar] [CrossRef]
- Martín, I.; Rodríguez, A.; Sánchez-Montero, L.; Padilla, P.; Córdoba, J.J. Effect of the dry-cured fermented sausage “salchichón” processing with a selected Lactobacillus sakei in Listeria monocytogenes and microbial population. Foods 2021, 10, 856. [Google Scholar] [CrossRef]
- Bungenstock, L.; Abdulmawjood, A.; Reich, F. Evaluation of antibacterial properties of lactic acid bacteria from traditionally and industrially produced fermented sausages from Germany. PLoS ONE 2020, 15, e0230345. [Google Scholar] [CrossRef]
- De Carvalho, A.A.T.; De Paula, R.A.; Mantovani, H.C.; De Moraes, C.A. Inhibition of Listeria monocytogenes by a lactic acid bacterium isolated from Italian salami. Food Microbiol. 2006, 23, 213–219. [Google Scholar] [CrossRef]
- Pedonese, F.; Torracca, B.; Mancini, S.; Pisano, S.; Turchi, B.; Cerri, D.; Nuvoloni, R. Effect of a Lactobacillus sakei and Staphylococcus xylosus protective culture on Listeria monocytogenes growth and quality traits of Italian fresh sausage (salsiccia) stored at abusive temperature. Ital. J. Anim. Sci. 2020, 19, 1363–1374. [Google Scholar] [CrossRef]
- dos Cruxen, C.E.S.; Funck, G.D.; Haubert, L.; da Dannenberg, G.S.; de Marques, J.L.; Chaves, F.C.; da Silva, W.P.; Fiorentini, Â.M. Selection of native bacterial starter culture in the production of fermented meat sausages: Application potential, safety aspects, and emerging technologies. Food Res. Int. 2019, 122, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Kasra-Kermanshahi, R.; Mobarak-Qamsari, E. Inhibition effect of lactic acid bacteria against food born pathogen, Listeria monocytogenes. Appl. Food Biotechnol. 2015, 2, 11–19. [Google Scholar] [CrossRef]
- Vieco-Saiz, N.; Belguesmia, Y.; Raspoet, R.; Auclair, E.; Gancel, F.; Kempf, I.; Drider, D. Benefits and inputs from lactic acid bacteria and their bacteriocins as alternatives to antibiotic growth promoters during food-animal production. Front. Microbiol. 2019, 10, 57. [Google Scholar] [CrossRef] [Green Version]
- Leroy, F.; De Vuyst, L. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci. Technol. 2004, 15, 67–78. [Google Scholar] [CrossRef]
- Singh, V.P. Recent approaches in food bio-preservation—A review. Open Vet. J. 2018, 8, 104–111. [Google Scholar] [CrossRef] [Green Version]
- Simons, A.; Alhanout, K.; Duval, R.E. Bacteriocins, antimicrobial peptides from bacterial origin: Overview of their biology and their impact against multidrug-resistant bacteria. Microorganisms 2020, 8, 639. [Google Scholar] [CrossRef] [PubMed]
- Mokoena, M.P.; Omatola, C.A.; Olaniran, A.O. Applications of lactic acid bacteria and their bacteriocins against food spoilage microorganisms and foodborne pathogens. Molecules 2021, 26, 7055. [Google Scholar] [CrossRef] [PubMed]
- Russo, P.; Arena, M.P.; Fiocco, D.; Capozzi, V.; Drider, D.; Spano, G. Lactobacillus plantarum with broad antifungal activity: A promising approach to increase safety and shelf-life of cereal-based products. Int. J. Food Microbiol. 2017, 247, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Dittoe, D.K.; Ricke, S.C.; Kiess, A.S. Organic acids and potential for modifying the avian gastrointestinal tract and reducing pathogens and disease. Front. Vet. Sci. 2018, 5, 1–12. [Google Scholar] [CrossRef]
- Ricke, S.C. Perspectives on the use of organic acids and short chain fatty acids as antimicrobials. Poult. Sci. 2003, 82, 632–639. [Google Scholar] [CrossRef]
- Nair, S.M.; Amalaradjou, M.A.; Venkitanarayanan, K. Antivirulence properties of probiotics in combating microbial pathogenesis. Adv. Appl. Microbiol. 2017, 98, 1–29. [Google Scholar] [CrossRef]
- Wang, C.; Chang, T.; Yang, H.; Cui, M. Antibacterial mechanism of lactic acid on physiological and morphological properties of Salmonella Enteritidis, Escherichia coli and Listeria monocytogenes. Food Control 2015, 47, 231–236. [Google Scholar] [CrossRef]
- Wemmenhove, E.; van Valenberg, H.J.F.; van Hooijdonk, A.C.M.; Wells-Bennik, M.H.J.; Zwietering, M.H. Factors that inhibit growth of Listeria monocytogenes in nature-ripened Gouda cheese: A major role for undissociated lactic acid. Food Control 2018, 84, 413–418. [Google Scholar] [CrossRef]
- Mokoena, M.P.; Mutanda, T.; Olaniran, O. Perspectives on the probiotic potential of lactic acid bacteria from African traditional fermented foods and beverages Mduduzi. Food Nutr. Res. 2016, 60, 29630. [Google Scholar] [CrossRef] [Green Version]
- Zúñiga, M.; Pardo, I.; Ferrer, S. An improved medium for distinguishing between homofermentative and heterofermentative lactic acid bacteria. Int. J. Food Microbiol. 1993, 18, 37–42. [Google Scholar] [CrossRef]
- Barker, C.; Park, S.F. Sensitization of Listeria monocytogenes to low pH, organic acids, and osmotic stress by ethanol. Appl. Environ. Microbiol. 2001, 67, 1594–1600. [Google Scholar] [CrossRef] [Green Version]
- Ghalfi, H.; Thonart, P.; Benkerroum, N. Inhibitory activity of Lactobacillus curvatus CWBI-B28 against Listeria monocytogenes and ST2-verotoxin producing Escherichia coli O157. Afr. J. Biotechnol. 2006, 5, 2303–2306. [Google Scholar] [CrossRef]
- Langa, S.; Martín-Cabrejas, I.; Montiel, R.; Landete, J.M.; Medina, M.; Arqués, J.L. Short communication: Combined antimicrobial activity of reuterin and diacetyl against foodborne pathogens. J. Dairy Sci. 2014, 97, 6116–6121. [Google Scholar] [CrossRef] [Green Version]
- Yap, P.-C.; MatRahim, N.-A.; AbuBakar, S.; Lee, H.Y. Antilisterial potential of lactic acid bacteria in eliminating Listeria monocytogenes in host and ready-to-eat food application. Microbiol. Res. 2021, 12, 234–257. [Google Scholar] [CrossRef]
- Saraoui, T.; Fall, P.A.; Leroi, F.; Antignac, J.P.; Chéreau, S.; Pilet, M.F. Inhibition mechanism of Listeria monocytogenes by a bioprotective bacteria Lactococcus piscium CNCM I-4031. Food Microbiol. 2016, 53, 70–78. [Google Scholar] [CrossRef] [Green Version]
- Zilelidou, E.; Rychli, K.; Manthou, E.; Ciolacu, L.; Wagner, M.; Skandamis, P.N. Highly invasive Listeria monocytogenes strains have growth and invasion advantages in strain competition. PLoS ONE 2015, 10, e0141617. [Google Scholar] [CrossRef] [Green Version]
- Bermudez-Brito, M.; Plaza-Díaz, J.; Muñoz-Quezada, S.; Gómez-Llorente, C.; Gil, A. Probiotic mechanisms of action. Ann. Nutr. Metab. 2012, 61, 160–174. [Google Scholar] [CrossRef]
- Herich, R.; Levkut, M. Lactic acid bacteria, probiotics and immune system. Vet. Med. 2002, 47, 169–180. [Google Scholar] [CrossRef] [Green Version]
- Corr, S.C.; Gahan, C.G.M.; Hill, C. Impact of selected Lactobacillus and Bifidobacterium species on Listeria monocytogenes infection and the mucosal immune response. FEMS Immunol. Med. Microbiol. 2007, 50, 380–388. [Google Scholar] [CrossRef] [Green Version]
- Ndahetuye, J.B.; Koo, O.K.; O’Bryan, C.A.; Ricke, S.C.; Crandall, P.G. Role of lactic acid bacteria as a biosanitizer to prevent attachment of Listeria monocytogenes F6900 on deli slicer contact surfaces. J. Food Prot. 2012, 75, 1429–1436. [Google Scholar] [CrossRef]
- Pilchová, T.; Pilet, M.F.; Cappelier, J.M.; Pazlarová, J.; Tresse, O. Protective effect of Carnobacterium spp. against Listeria monocytogenes during host cell invasion using in vitro HT29 model. Front. Cell. Infect. Microbiol. 2016, 6, 88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gómez, N.C.; Ramiro, J.M.P.; Quecan, B.X.V.; de Melo Franco, B.D.G. Use of potential probiotic lactic acid bacteria (LAB) biofilms for the control of Listeria monocytogenes, Salmonella Typhimurium, and Escherichia coli O157: H7 biofilms formation. Front. Microbiol. 2016, 7, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dutra, V.; Silva, A.C.; Cabrita, P.; Peres, C.; Malcata, X.; Brito, L. Lactobacillus plantarum LB95 impairs the virulence potential of Gram-positive and Gram-negative food-borne pathogens in HT-29 and vero cell cultures. J. Med. Microbiol. 2016, 65, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, A.; Upadhyaya, I.; Mooyottu, S.; Venkitanarayanan, K. Eugenol in combination with lactic acid bacteria attenuates Listeria monocytogenes virulence in vitro and in invertebrate model Galleria mellonella. J. Med. Microbiol. 2016, 65, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Garriga, M.; Rubio, R.; Aymerich, T.; Ruas-Madiedo, P. Potentially probiotic and bioprotective lactic acid bacteria starter cultures antagonise the Listeria monocytogenes adhesion to HT29 colonocyte-like cells. Benef. Microbes 2015, 6, 337–343. [Google Scholar] [CrossRef] [Green Version]
- Winkelströter, L.K.; De Martinis, E.C.P. Effect of bacteriocins and conditions that mimic food and digestive tract on biofilm formation, in vitro invasion of eukaryotic cells and Internalin gene expression by Listeria monocytogenes. Probiotics Antimicrob. Proteins 2013, 5, 153–164. [Google Scholar] [CrossRef]
- De Vuyst, L.; Leroy, F. Bacteriocins from lactic acid bacteria: Production, purification, and food applications. J. Mol. Microbiol. Biotechnol. 2007, 13, 194–199. [Google Scholar] [CrossRef]
- Pessione, E. Lactic acid bacteria contribution to gut microbiota complexity: Lights and shadows. Front. Cell. Infect. Microbiol. 2012, 2, 86. [Google Scholar] [CrossRef] [Green Version]
- Vogel, R.F.; Pohle, B.S.; Tichaczek, P.S.; Hammes, W.P. The competitive advantage of Lactobacillus curvatus LTH 1174 in sausage fermentations is caused by formation of curvacin A. Syst. Appl. Microbiol. 1993, 16, 457–462. [Google Scholar] [CrossRef]
- Cheigh, C.I.; Pyun, Y.R. Nisin biosynthesis and its properties. Biotechnol. Lett. 2005, 27, 1641–1648. [Google Scholar] [CrossRef]
- Klaenhammer, T.R. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 1993, 12, 39–85. [Google Scholar] [CrossRef]
- Deegan, L.H.; Cotter, P.D.; Hill, C.; Ross, P. Bacteriocins: Biological tools for bio-preservation and shelf-life extension. Int. Dairy J. 2006, 16, 1058–1071. [Google Scholar] [CrossRef]
- Férir, G.; Petrova, M.I.; Andrei, G.; Huskens, D.; Hoorelbeke, B.; Snoeck, R.; Vanderleyden, J.; Balzarini, J.; Bartoschek, S.; Brönstrup, M.; et al. The lantibiotic peptide labyrinthopeptin A1 demonstrates broad anti-HIV and anti-HSV activity with potential for microbicidal applications. PLoS ONE 2013, 8, e64010. [Google Scholar] [CrossRef] [Green Version]
- Kawulka, K.E.; Sprules, T.; Diaper, C.M.; Whittal, R.M.; McKay, R.T.; Mercier, P.; Zuber, P.; Vederas, J.C. Structure of subtilosin A, a cyclic antimicrobial peptide from Bacillus subtilis with unusual sulfur to α-carbon cross-links: Formation and reduction of α-Thio-α-Amino acid derivatives. Biochemistry 2004, 43, 3385–3395. [Google Scholar] [CrossRef]
- Gao, Y.; Li, D.; Liu, S.; Zhang, L. Garviecin LG34, a novel bacteriocin produced by Lactococcus garvieae isolated from traditional Chinese fermented cucumber. Food Control 2015, 50, 896–900. [Google Scholar] [CrossRef]
- Fimland, G.; Jack, R.; Jung, G.; Nes, I.F.; Nissen-Meyer, J. The bactericidal activity of pediocin PA-1 is specifically inhibited by a 15-mer fragment that spans the bacteriocin from the Center toward the C terminus. Appl. Environ. Microbiol. 1998, 64, 5057–5060. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Tian, F.; Li, S.; Xie, Y.; Zhang, H.; Chen, W. Cloning and heterologous expression of a bacteriocin sakacin P from Lactobacillus sakei in Escherichia coli. Appl. Microbiol. Biotechnol. 2012, 94, 1061–1068. [Google Scholar] [CrossRef]
- Trinetta, V.; Morleo, A.; Sessa, F.; Iametti, S.; Bonomi, F.; Ferranti, P. Purified sakacin A shows a dual mechanism of action against Listeria spp: Proton motive force dissipation and cell wall breakdown. FEMS Microbiol. Lett. 2012, 334, 143–149. [Google Scholar] [CrossRef] [Green Version]
- Ceuppens, S.; De Coninck, D.; Bottledoorn, N.; Van Nieuwerburgh, F.; Uyttendaele, M. Microbial community profiling of fresh basil and pitfalls in taxonomic assignment of enterobacterial pathogenic species based upon 16S rRNA amplicon sequencing. Int. J. Food Microbiol. 2017, 257, 148–156. [Google Scholar] [CrossRef]
- Ekblad, B.; Nissen-Meyer, J.; Kristensen, T. Whole-genome sequencing of mutants with increased resistance against the two-peptide bacteriocin plantaricin JK reveals a putative receptor and potential docking site. PLoS ONE 2017, 12, e0185279. [Google Scholar] [CrossRef] [Green Version]
- Ekblad, B.; Kyriakou, P.K.; Oppegård, C.; Nissen-Meyer, J.; Kaznessis, Y.N.; Kristiansen, P.E. Structure-function analysis of the two-peptide bacteriocin plantaricin EF. Biochemistry 2016, 55, 5106–5116. [Google Scholar] [CrossRef] [PubMed]
- Oppegård, C.; Rogne, P.; Kristiansen, P.E.; Nissen-Meyer, J. Structure analysis of the two-peptide bacteriocin lactococcin G by introducing D-amino acid residues. Microbiology 2010, 156, 1883–1889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gálvez, A.; Maqueda, M.; Martínez-Bueno, M.; Valdivia, E. Bactericidal and bacteriolytic action of peptide antibiotic AS-48 against gram-positive and gram-negative bacteria and other organisms. Res. Microbiol. 1989, 140, 57–68. [Google Scholar] [CrossRef]
- Pandey, N.; Malik, R.K.; Kaushik, J.K.; Singroha, G. Gassericin A: A circular bacteriocin produced by Lactic acid bacteria Lactobacillus gasseri. World J. Microbiol. Biotechnol. 2013, 29, 1977–1987. [Google Scholar] [CrossRef]
- Chehimi, S.; Pons, A.M.; Sablé, S.; Hajlaoui, M.R.; Limam, F. Mode of action of thuricin S, a new class IId bacteriocin from Bacillus thuringiensis. Can. J. Microbiol. 2010, 56, 162–167. [Google Scholar] [CrossRef]
- O’Shea, E.F.; O’Connor, P.M.; O’Sullivan, O.; Cotter, P.D.; Ross, R.P.; Hill, C. Bactofencin A, a new type of cationic bacteriocin with Unusual Immunity. mBio 2013, 4, e00498-13. [Google Scholar] [CrossRef] [Green Version]
- Schindler, C.A.; Schuhardt, V.T. Lysostaphin; a new bacteriolytic agent for the Staphylococcus. Proc. Natl. Acad. Sci. USA 1964, 51, 414–421. [Google Scholar] [CrossRef] [Green Version]
- Vaughan, E.E.; Daly, C.; Fitzgerald, G.F. Identification and characterization of helveticin V-1829, a bacteriocin produced by Lactobacillus helveticus 1829. J. Appl. Bacteriol. 1992, 73, 299–308. [Google Scholar] [CrossRef]
- Maky, M.A.; Ishibashi, N.; Zendo, T.; Perez, R.H.; Doud, J.R.; Karmi, M.; Sonomoto, K. Enterocin F4-9, a novel O-linked glycosylated bacteriocin. Appl. Environ. Microbiol. 2015, 81, 4819–4826. [Google Scholar] [CrossRef] [Green Version]
- Breukink, E.; Van Heusden, H.E.; Vollmerhaus, P.J.; Swiezewska, E.; Brunner, L.; Walker, S.; Heck, A.J.R.; De Kruijff, B. Lipid II is an intrinsic component of the pore induced by nisin in bacterial membranes. J. Biol. Chem. 2003, 278, 19898–19903. [Google Scholar] [CrossRef] [Green Version]
- Breukink, E.; De Kruijff, B. The lantibiotic nisin, a special case or not? Biochim. Biophys. Acta—Biomembr. 1999, 1462, 223–234. [Google Scholar] [CrossRef] [Green Version]
- European Commission. Commission Regulation (EU) No 1129/2011 of 11 November 2011 amending Annex II to Regulation (EC) No 1333/2008 of the European Parliament and of the Council by establishing a Union list of food additives. Off. J. Eur. Union 2011, L295, 1–177. [Google Scholar] [CrossRef]
- Zhu, L.; Zeng, J.; Wang, C.; Wang, J. Structural basis of pore formation in the mannose phosphotransferase system (man-PTS) by pediocin PA-1. Appl. Environ. Microbiol. 2021, 88, e0199221. [Google Scholar] [CrossRef]
- Schillinger, U.; Kaya, M.; Lücke, F.-K. Behaviour of Listeria monocytogenes in meat and its control by a bacteriocin-producing strain of Lactobacillus sake. J. Appl. Bacteriol. 1991, 70, 473–478. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, H.; Kuo, C.; Chen, Y.; Ho, S.; Yanagida, F. Leucocin C-607, a Novel Bacteriocin from the multiple-bacteriocin-producing Leuconostoc pseudomesenteroides 607 isolated from persimmon. Probiotics Antimicrob. Proteins 2018, 10, 148–156. [Google Scholar] [CrossRef]
- Liu, G.; Ren, L.; Song, Z.; Wang, C.; Sun, B. Purification and characteristics of bifidocin A, a novel bacteriocin produced by Bifidobacterium animals BB04 from centenarians’ intestine. Food Control 2015, 50, 889–895. [Google Scholar] [CrossRef]
- Wang, Y.; Qin, Y.; Zhang, Y.; Wu, R.; Li, P. Antibacterial mechanism of plantaricin LPL-1, a novel class IIa bacteriocin against Listeria monocytogenes. Food Control 2019, 97, 87–93. [Google Scholar] [CrossRef]
- Diep, D.B.; Skaugen, M.; Salehian, Z.; Holo, H.; Nes, I.F. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc. Natl. Acad. Sci. USA 2007, 104, 2384–2389. [Google Scholar] [CrossRef] [Green Version]
- Jeckelmann, J.M.; Erni, B. The mannose phosphotransferase system (Man-PTS)—Mannose transporter and receptor for bacteriocins and bacteriophages. Biochim. Biophys. Acta—Biomembr. 2020, 1862, 183412. [Google Scholar] [CrossRef]
- Liu, X.; Zeng, J.; Huang, K.; Wang, J. Structure of the mannose transporter of the bacterial phosphotransferase system. Cell Res. 2019, 29, 680–682. [Google Scholar] [CrossRef]
- Chikindas, M.L.; Garcia-Garcera, M.J.; Driessen, A.J.M.; Ledeboer, A.M.; Nissen-Meyer, J.; Nes, I.F.; Abee, T.; Konings, W.N.; Venema, G. Pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC1.0, forms hydrophilic pores in the cytoplasmic membrane of target cells. Appl. Environ. Microbiol. 1993, 59, 3577–3584. [Google Scholar] [CrossRef] [Green Version]
- Moll, G.N.; Konings, W.N.; Driessen, A.J.M. Bacteriocins: Mechanism of membrane insertion and pore formation. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 1999, 76, 185–198. [Google Scholar] [CrossRef]
- Rashid, R.; Veleba, M.; Kline, K.A. Focal targeting of the bacterial envelope by antimicrobial peptides. Front. Cell Dev. Biol. 2016, 4, 1–13. [Google Scholar] [CrossRef]
- Mazzotta, A.S.; Montville, T.J. Nisin induces changes in membrane fatty acid composition of Listeria monocytogenes nisin-resistant strains at 10 °C and 30 °C. J. Appl. Microbiol. 1997, 82, 32–38. [Google Scholar] [CrossRef]
- Demel, R.A.; Peelen, T.; Siezen, R.J.; De Kruijff, B.; Kuipers, O.P. Nisin Z, mutant nisin Z and lacticin 481 interactions with anionic lipids correlate with antimicrobial activity: A monolayer study. Eur. J. Biochem. 1996, 235, 267–274. [Google Scholar] [CrossRef] [Green Version]
- Van Kraaij, C.; Breukink, E.; Noordermeer, M.A.; Demel, R.A.; Siezen, R.J.; Kuipers, O.P.; De Kruijff, B. Pore formation by nisin involves translocation of its C-terminal part across the membrane. Biochemistry 1998, 37, 16033–16040. [Google Scholar] [CrossRef] [Green Version]
- Gravesen, A.; Ramnath, M.; Rechinger, K.B.; Andersen, N.; Jänsch, L.; Héchard, Y.; Hastings, J.W.; Knøchel, S. High-level resistance to class IIa bacteriocins is associated with one general mechanism in Listeria monocytogenes. Microbiology 2002, 148, 2361–2369. [Google Scholar] [CrossRef] [Green Version]
- Opsata, M.; Nes, I.F.; Holo, H. Class IIa bacteriocin resistance in Enterococcus faecalis V583: The mannose PTS operon mediates global transcriptional responses. BMC Microbiol. 2010, 10, 224. [Google Scholar] [CrossRef] [Green Version]
- Kumariya, R.; Garsa, A.K.; Rajput, Y.S.; Sood, S.K.; Akhtar, N.; Patel, S. Bacteriocins: Classification, synthesis, mechanism of action and resistance development in food spoilage causing bacteria. Microb. Pathog. 2019, 128, 171–177. [Google Scholar] [CrossRef]
- Barbieri, F.; Montanari, C.; Gardini, F.; Tabanelli, G. Biogenic amine production by lactic acid bacteria: A review. Foods 2019, 8, 17. [Google Scholar] [CrossRef] [Green Version]
- Biscola, V.; Choiset, Y.; Rabesona, H.; Chobert, J.M.; Haertlé, T.; Franco, B.D.G.M. Brazilian artisanal ripened cheeses as sources of proteolytic lactic acid bacteria capable of reducing cow milk allergy. J. Appl. Microbiol. 2018, 125, 564–574. [Google Scholar] [CrossRef] [PubMed]
- Bintsis, T. Lactic acid bacteria as starter cultures: An update in their metabolism and genetics. AIMS Microbiol. 2018, 4, 665–684. [Google Scholar] [CrossRef] [PubMed]
- Marshall, B.M.; Levy, S.B. Food animals and antimicrobials: Impacts on human health. Clin. Microbiol. Rev. 2011, 24, 718–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, M.; Barbosa, J.; Albano, H.; Teixeira, P. Bacteriocinogenic activity of Leuconostoc lactis RK18 isolates from fermented food. In Fermented Foods; Oliver, K., Ed.; Nova Science Publishers, Inc.: New York, NY, USA, 2020; Chapter 3; p. 159. [Google Scholar]
- Mariam, S.H.; Zegeye, N.; Tariku, T.; Andargie, E.; Endalafer, N.; Aseffa, A. Potential of cell-free supernatants from cultures of selected lactic acid bacteria and yeast obtained from local fermented foods as inhibitors of Listeria monocytogenes, Salmonella spp. and Staphylococcus aureus. BMC Res. Notes 2014, 7, 606. [Google Scholar] [CrossRef] [Green Version]
- Schägger, H.; von Jagow, G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987, 166, 368–379. [Google Scholar] [CrossRef]
- Barbosa, J.; Albano, H.; Silva, B.; Almeida, M.H.; Nogueira, T.; Teixeira, P. Characterization of a Lactiplantibacillus plantarum r23 isolated from arugula by whole-genome sequencing and its bacteriocin production ability. Int. J. Environ. Res. Public Health 2021, 18, 5515. [Google Scholar] [CrossRef]
- Abrams, D.; Barbosa, J.; Albano, H.; Silva, J.; Gibbs, P.A.; Teixeira, P. Characterization of bacPPK34 a bacteriocin produced by Pediococcus pentosaceus strain K34 isolated from “Alheira”. Food Control 2011, 22, 940–946. [Google Scholar] [CrossRef]
- Nebbia, S.; Lamberti, C.; Lo Bianco, G.; Cirrincione, S.; Laroute, V.; Cocaign-Bousquet, M.; Cavallarin, L.; Giuffrida, M.G.; Pessione, E. Antimicrobial potential of food lactic acid bacteria: Bioactive peptide decrypting from caseins and bacteriocin production. Microorganisms 2021, 9, 65. [Google Scholar] [CrossRef]
- Azizi, F.; Habibi Najafi, M.B.; Dovom, M.R.E. The biodiversity of Lactobacillus spp. from Iranian raw milk Motal cheese and antibacterial evaluation based on bacteriocin-encoding genes. AMB Express 2017, 7, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Ho, V.T.T.; Lo, R.; Bansal, N.; Turner, M.S. Characterisation of Lactococcus lactis isolates from herbs, fruits and vegetables for use as biopreservatives against Listeria monocytogenes in cheese. Food Control 2018, 85, 472–483. [Google Scholar] [CrossRef] [Green Version]
- Bhatti, M.; Veeramachaneni, A.; Shelef, L.A. Factors affecting the antilisterial effects of nisin in milk. Int. J. Food Microbiol. 2004, 97, 215–219. [Google Scholar] [CrossRef]
- Zapico, P.; Medina, M.; Gaya, P.; Nuñez, M. Synergistic effect of nisin and the lactoperoxidase system on Listeria monocytogenes in skim milk. Int. J. Food Microbiol. 1998, 40, 35–42. [Google Scholar] [CrossRef]
- García-Díez, J.; Patarata, L. Influence of salt level, starter culture, fermentable carbohydrates, and temperature on the behaviour of L. monocytogenes in sliced chouriço during storage. Acta Aliment. 2017, 46, 206–213. [Google Scholar] [CrossRef] [Green Version]
- Ortiz, S.; López, V.; Garriga, M.; Martínez-Suárez, J.V. Antilisterial effect of two bioprotective cultures in a model system of Iberian chorizo fermentation. Int. J. Food Sci. Technol. 2014, 49, 753–758. [Google Scholar] [CrossRef]
- Vaz-Velho, M.; Jácome, S.; Noronha, L.; Todorov, S.; Fonseca, S.; Pinheiro, R.; Morais, A.; Silva, J.; Teixeira, P. Comparison of antilisterial effects of two strains of lactic acid bacteria during processing and storage of a portuguese salami-like product alheira. Chem. Eng. Trans. 2013, 32, 1807–1812. [Google Scholar] [CrossRef]
- Kamiloglu, A.; Kaban, G.; Kaya, M. Effects of autochthonous Lactobacillus plantarum strains on Listeria monocytogenes in sucuk during ripening. J. Food Saf. 2019, 39, 1–8. [Google Scholar] [CrossRef]
- Zanette, C.M.; Dalla Santa, O.R.; Bersot, L.S. Effect of Lactobacillus plantarum starter cultures on the behavior of Listeria monocytogenes during sausage maturation. Int. Food Res. J. 2015, 22, 844–848. [Google Scholar]
- Castellano, P.; Peña, N.; Ibarreche, M.P.; Carduza, F.; Soteras, T.; Vignolo, G. Antilisterial efficacy of Lactobacillus bacteriocins and organic acids on frankfurters. Impact on sensory characteristics. J. Food Sci. Technol. 2018, 55, 689–697. [Google Scholar] [CrossRef]
- Nikodinoska, I.; Baffoni, L.; Di Gioia, D.; Manso, B.; García-Sánchez, L.; Melero, B.; Rovira, J. Protective cultures against foodborne pathogens in a nitrite reduced fermented meat product. LWT 2019, 101, 293–299. [Google Scholar] [CrossRef]
- Macieira, A.; Barros, D.; Vaz-Velho, M.; Pinheiro, R.; Fonseca, S.; Albano, H.; Teixeira, P. Effects of Lactobacillus plantarum bacteriocinogenic culture on physicochemical, microbiological, and sensorial characteristics of “Chouriço Vinha d’Alhos”, A traditional Portuguese Sausage. J. Food Qual. Hazards Control 2018, 5, 118–127. [Google Scholar] [CrossRef]
- Khalili Sadaghiani, S.; Aliakbarlu, J.; Tajik, H.; Mahmoudian, A. Anti-Listeria activity and shelf life extension effects of Lactobacillus along with garlic extract in ground beef. J. Food Saf. 2019, 39, 1–8. [Google Scholar] [CrossRef]
- Cosansu, S.; Geornaras, I.; Ayhan, K.; Sofos, J.N. Control of Listeria monocytogenes by bacteriocin-producing Pediococcus acidilactici 13 and its antimicrobial substance in a dry fermented sausage sucuk and in turkey breast. J. Food Nutr. Res. 2010, 49, 206–214. [Google Scholar]
- Olaoye, O.A.; Onilude, A.A. Investigation on the potential application of biological agents in the extension of shelf life of fresh beef in Nigeria. World J. Microbiol. Biotechnol. 2010, 26, 1445–1454. [Google Scholar] [CrossRef]
- Orihuel, A.; Bonacina, J.; Vildoza, M.J.; Bru, E.; Vignolo, G.; Saavedra, L.; Fadda, S. Biocontrol of Listeria monocytogenes in a meat model using a combination of a bacteriocinogenic strain with curing additives. Food Res. Int. 2018, 107, 289–296. [Google Scholar] [CrossRef]
- Trinetta, V.; Floros, J.D.; Cutter, C.N. Sakacin a-containing pullulan film: An active packaging system to control epidemic clones of Listeria monocytogenes in ready-to-eat foods. J. Food Saf. 2010, 30, 366–381. [Google Scholar] [CrossRef]
- Ruiz, A.; Williams, S.K.; Djeri, N.; Hinton, A.; Rodrick, G.E. Nisin affects the growth of Listeria monocytogenes on ready-to-eat turkey ham stored at four degrees Celsius for sixty-three days. Poult. Sci. 2010, 89, 353–358. [Google Scholar] [CrossRef]
- Balay, D.R.; Dangeti, R.V.; Kaur, K.; McMullen, L.M. Purification of leucocin A for use on wieners to inhibit Listeria monocytogenes in the presence of spoilage organisms. Int. J. Food Microbiol. 2017, 255, 25–31. [Google Scholar] [CrossRef]
- Kondrotiene, K.; Kasnauskyte, N.; Serniene, L.; Gölz, G.; Alter, T.; Kaskoniene, V.; Maruska, A.S.; Malakauskas, M. Characterization and application of newly isolated nisin producing Lactococcus lactis strains for control of Listeria monocytogenes growth in fresh cheese. LWT—Food Sci. Technol. 2018, 87, 507–514. [Google Scholar] [CrossRef]
- Ivanovic, M.; Mirkovic, N.; Mirkovic, M.; Miocinovic, J.; Radulovic, A.; Knudsen, T.S.; Radulovic, Z. Autochthonous Enterococcus durans PFMI565 and Lactococcus lactis subsp. lactis BGBU1–4 in bio-control of Listeria monocytogenes in ultrafiltered cheese. Foods 2021, 10, 1448. [Google Scholar] [CrossRef]
- Mills, S.; Serrano, L.M.; Griffin, C.; O’Connor, P.M.; Schaad, G.; Bruining, C.; Hill, C.; Ross, R.P.; Meijer, W.C. Inhibitory activity of Lactobacillus plantarum LMG P-26358 against Listeria innocua when used as an adjunct starter in the manufacture of cheese. Microb. Cell Fact. 2011, 10, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Hernández, D.; Cardell, E.; Zárate, V. Antimicrobial activity of lactic acid bacteria isolated from Tenerife cheese: Initial characterization of plantaricin TF711, a bacteriocin like substance produced by Lactobacillus plantarum TF711. J. Appl. Microbiol. 2005, 99, 77–84. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, L.; O’Connor, E.; Ross, R.; Hill, C. Evaluation of live-culture producing lacticin 3147 as a treatment for the control of Listeria monocytogenes on the surface of smear-ripened cheese. J. Appl. Microbiol. 2006, 100, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Falardeau, J.; Trmčić, A.; Wang, S. The occurrence, growth, and biocontrol of Listeria monocytogenes in fresh and surface-ripened soft and semisoft cheeses. Compr. Rev. Food Sci. Food Saf. 2021, 20, 4019–4048. [Google Scholar] [CrossRef] [PubMed]
- Lourenço, A.; Kamnetz, M.B.; Gadotti, C.; Diez-Gonzalez, F. Antimicrobial treatments to control Listeria monocytogenes in queso fresco. Food Microbiol. 2017, 64, 47–55. [Google Scholar] [CrossRef]
- de Pimentel-Filho, N.J.; Mantovani, H.C.; de Carvalho, A.F.; Dias, R.S.; Vanetti, M.C.D. Efficacy of bovicin HC5 and nisin combination against Listeria monocytogenes and Staphylococcus aureus in fresh cheese. Int. J. Food Sci. Technol. 2014, 49, 416–422. [Google Scholar] [CrossRef]
- Dal Bello, B.; Cocolin, L.; Zeppa, G.; Field, D.; Cotter, P.D.; Hill, C. Technological characterization of bacteriocin producing Lactococcus lactis strains employed to control Listeria monocytogenes in Cottage cheese. Int. J. Food Microbiol. 2012, 153, 58–65. [Google Scholar] [CrossRef]
- Loessner, M.; Guenther, S.; Steffan, S.; Scherer, S. A pediocin-producing Lactobacillus plantarum strain inhibits Listeria monocytogenes in a multispecies cheese surface microbial ripening consortium. Appl. Environ. Microbiol. 2003, 69, 1854–1857. [Google Scholar] [CrossRef] [Green Version]
- Ribeiro, S.C.; O’Connor, P.M.; Ross, R.P.; Stanton, C.; Silva, C.C.G. An anti-listerial Lactococcus lactis strain isolated from Azorean Pico cheese produces lacticin 481. Int. Dairy J. 2016, 63, 18–28. [Google Scholar] [CrossRef]
- Ribeiro, S.C.; Ross, R.P.; Stanton, C.; Silva, C.C.G. Characterization and application of antilisterial enterocins on model fresh cheese. J. Food Prot. 2017, 80, 1303–1316. [Google Scholar] [CrossRef]
- Possas, A.; Bonilla-Luque, O.M.; Valero, A. From cheese-making to consumption: Exploring the microbial safety of cheeses through predictive microbiology models. Foods 2021, 10, 335. [Google Scholar] [CrossRef]
- Arqués, J.L.; Rodríguez, E.; Langa, S.; Landete, J.M.; Medina, M. Antimicrobial activity of lactic acid bacteria in dairy products and gut: Effect on pathogens. Biomed Res. Int. 2015, 2015, 584183. [Google Scholar] [CrossRef]
- Morandi, S.; Silvetti, T.; Vezzini, V.; Morozzo, E.; Brasca, M. How we can improve the antimicrobial performances of lactic acid bacteria? A new strategy to control Listeria monocytogenes in Gorgonzola cheese. Food Microbiol. 2020, 90, 103488. [Google Scholar] [CrossRef]
- Contessa, C.R.; De Souza, N.B.; Batt, G.; De Moura, C.M.; Silveira, G.; Moraes, C.C. Development of active packaging based on agar-agar incorporated with bacteriocin of Lactobacillus sakei. Biomolecules 2021, 11, 1869. [Google Scholar] [CrossRef]

| Bifidobacterium adolescentis | Lactobacillus delbruechkii | Ligilactobacillus animalis |
| Bifidobacterium animalis | Lactobacillus gallinarum | Ligilactobacillus aviaries |
| Bifidobacterium bifidum | Lactobacillus gasseri | Ligilactobacillus salivarius |
| Bifidobacterium breve | Lactobacillus helveticus | Liminosilactobacillus fermentum |
| Bifidobacterium longum | Lactobacillus johnsonii | Liminosilactobacillus mucosae |
| Carnobacterium divergens | Lactobacillus kefiranofaciens | Liminosilactobacillus panis |
| Companilactobacillus alimentarius | Lactococcus lactis | Liminosilactobacillus pontis |
| Companilactobacillus farciminis | Lapidilactobacillus dextrinicus | Liminosilactobacillus reuteri |
| Corynebacterium ammoniagenes | Latilactobacillus curvatus | Loigolactobacillus coryniformis |
| Corynebacterium glutamicum | Latilactobacillu sakei | Microbacterium imperial |
| Fructilactobacillus sanfranciscensis | Lentilactobacillus buchneri | Oenococcus oeni |
| Lacticaseibacillus casei | Lentilactobacillus diolivorans | Pasteuria nishizawae |
| Lacticaseibacillus paracasei | Lentilactobacillus hilgardii | Pediococcus acidilactici |
| Lacticaseibacillus rhamnosus | Lentilactobacillus kefiri | Pediococcus parvulus |
| Lactiplantibacillus pentosus | Lentilactobacillus parafarraginis | Pediococcus pentosaceus |
| Lactiplantibacillus plantarum | Lentilactobacillus paraplantarum | Propionibacterium acidipropionici |
| Lactobacillus acidophilus | Leuconostoc citreum | Propionibacterium freudenreichii |
| Lactobacillus amylolyticus | Leuconostoc lactis | Secundilactobacillus collinoides |
| Lactobacillus amylovorus | Leuconostoc mesenteroides | Streptococcus thermophilus |
| Lactobacillus cellobiosus | Leuconostoc pseudomesenteroides | |
| Lactobacillus crispatus | Levilactobacillus brevis |
| Inhibitory Compound | Mechanism of Action | References |
|---|---|---|
| Lactic acid and other volatile acids | Disruption of cellular metabolism | [27] |
| Ethanol | Membrane fluidity and integrity | [28] |
| Hydrogen peroxide | Inactivation of essential biomolecules by superoxide anion chain reaction | [29] |
| Carbon dioxide | Anaerobic environment and/or inhibition of enzyme decarboxylation and/or disruption of the cell membrane | [30] |
| Diacetyl | Interference with arginine utilization | [29] |
| Bacteriocins | Disruption of cytoplasmic membrane | [27,31] |
| Class | Characteristics | Example | Producer | Reference |
|---|---|---|---|---|
| Ia | Lantibiotics (<5 KDa) | Nisin | Lactococcus lactis | [62] |
| Ib | Carbacyclic lantibiotics | Labyrinthopeptien A1 | Actinomadura nambiensis | [63] |
| Ic | Sactibiotics | Subtilosin A | Bacillus subtilis | [64] |
| IIa | Heat-stable peptides with N terminal- YGNGV | Pediocin PA-1, sakacins A and P, leucocin A, garviecin LG34 | Pediococcus pentosaceus, Pediococcus acidilactici, Lactilactobacillus sakei, Lactococcus garvieae | [65,66,67,68,69] |
| IIb | Two-peptide bacteriocins | Lactococcin G, plantaricin EF and JK | Lactiplantibacillus plantarum, Lactococcus spp. | [70,71,72] |
| IIc | Circular bacteriocins | Enterocin AS-48, gassericin A | Lactococcus gasseri, Enterococcus faecalis | [73,74] |
| IId | Single, linear, nonpediocin-like bacteriocins | Thuricin S, bactofencin A | Bacillus thuringensis, Ligilactobacillus salivarius | [75,76] |
| IIIa | Heat labile, >30 KDa with hydrolase activity | Lysostaphin | Staphylococcus. simulans biovar staphylolyticus | [77] |
| IIIb | Heat labile, >30 KDa without hydrolase activity | Helveticin | Lactobacillus helveticus | [78] |
| IV | Large complexes with carbohydrate or lipid moieties | Enterocin F4-9 | Enterococcus faecalis | [79] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martín, I.; Rodríguez, A.; Delgado, J.; Córdoba, J.J. Strategies for Biocontrol of Listeria monocytogenes Using Lactic Acid Bacteria and Their Metabolites in Ready-to-Eat Meat- and Dairy-Ripened Products. Foods 2022, 11, 542. https://doi.org/10.3390/foods11040542
Martín I, Rodríguez A, Delgado J, Córdoba JJ. Strategies for Biocontrol of Listeria monocytogenes Using Lactic Acid Bacteria and Their Metabolites in Ready-to-Eat Meat- and Dairy-Ripened Products. Foods. 2022; 11(4):542. https://doi.org/10.3390/foods11040542
Chicago/Turabian StyleMartín, Irene, Alicia Rodríguez, Josué Delgado, and Juan J. Córdoba. 2022. "Strategies for Biocontrol of Listeria monocytogenes Using Lactic Acid Bacteria and Their Metabolites in Ready-to-Eat Meat- and Dairy-Ripened Products" Foods 11, no. 4: 542. https://doi.org/10.3390/foods11040542
APA StyleMartín, I., Rodríguez, A., Delgado, J., & Córdoba, J. J. (2022). Strategies for Biocontrol of Listeria monocytogenes Using Lactic Acid Bacteria and Their Metabolites in Ready-to-Eat Meat- and Dairy-Ripened Products. Foods, 11(4), 542. https://doi.org/10.3390/foods11040542