ACE Inhibitory Peptides Derived from Muscovy Duck (Cairina moschata) Plasma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. MDPH Preparation
2.3. ACE Inhibition Activity
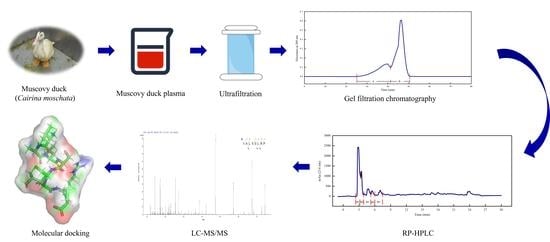
2.4. Peptide Separation and Purification
2.4.1. Ultrafiltration
2.4.2. Gel Filtration Chromatography
2.4.3. RP–HPLC
2.5. Identification of Peptide Sequences by LC–MS/MS
2.6. Prediction of ACE Inhibitory Activity
2.7. Peptide Synthesis
2.8. Molecular Docking
2.9. In Vitro Simulated Pepsin and Trypsin Digestion
2.10. In Silico Simulated Peptide Digestion
2.11. Statistical Analysis
3. Results and Discussion
3.1. Separation and Purification of MDPH
3.1.1. Ultrafiltration
3.1.2. Gel Filtration Chromatography
3.1.3. RP–HPLC
3.2. Identification of ACE–IPs
3.3. Validation of ACE Inhibitory Activity
3.4. Molecular Docking
3.5. In Vitro Simulated Pepsin and Trypsin Digestion and In Silico Simulated Peptide Digestion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lin, J.; Guan, L.; Ge, L.; Liu, G.; Bai, Y.; Liu, X. Nanopore-based full-length transcriptome sequencing of Muscovy duck (Cairina moschata) ovary. Poult. Sci. 2021, 100, 101246. [Google Scholar] [CrossRef]
- Zhu, Z.; Bassey, A.P.; Cao, Y.; Du, X.; Huang, T.; Cheng, Y.; Huang, M. Meat quality and flavor evaluation of Nanjing water boiled salted duck (NWSD) produced by different Muscovy duck (Cairina moschata) ingredients. Food Chem. 2022, 397, 133833. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Pan, X.; Cao, S.; Xu, F.; Lan, L.; Zhang, Y.; Lian, S.; Yan, M.; Li, A. iTRAQ-based quantitative proteomic analysis provides insights into strong broodiness in Muscovy duck (Cairina moschata) combined with metabolomics analysis. J. Proteom. 2019, 204, 103401. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Huang, J.; Zhu, Z.; Huang, M. Investigation of optimal conditions for production of antioxidant peptides from duck blood plasma: Response surface methodology. Poult. Sci. 2020, 99, 7159–7168. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Si, D.; Ahmad, B.; Li, Z.; Zhang, R. A novel antioxidative peptide derived from chicken blood corpuscle hydrolysate. Food Res. Int. 2018, 106, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Acharya, K.R.; Sturrock, E.D.; Riordan, J.F.; Ehlers, M.R. Ace revisited: A new target for structure-based drug design. Nat. Rev. Drug Discov. 2003, 2, 891–902. [Google Scholar] [CrossRef]
- Lee, S.; Qian, Z.; Kim, S. A novel angiotensin I converting enzyme inhibitory peptide from tuna frame protein hydrolysate and its antihypertensive effect in spontaneously hypertensive rats. Food Chem. 2010, 118, 96–102. [Google Scholar] [CrossRef]
- Xu, Z.; Wu, C.; Sun-Waterhouse, D.; Zhao, T.; Waterhouse, G.I.; Zhao, M.; Su, G. identification of post-digestion angiotensin-I converting enzyme (ACE) inhibitory peptides from soybean protein Isolate: Their production conditions and in silico molecular docking with ACE. Food Chem. 2021, 345, 128855. [Google Scholar] [CrossRef]
- Wu, J.; Liao, W.; Udenigwe, C.C. Revisiting the mechanisms of ACE inhibitory peptides from food proteins. Trends Food Sci. Technol. 2017, 69, 214–219. [Google Scholar] [CrossRef]
- Bhat, Z.F.; Kumar, S.; Bhat, H.F. Antihypertensive peptides of animal origin: A review. Crit. Rev. Food Sci. 2017, 57, 566–578. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, W.; Xue, J.; Yang, J.; Chen, X.; Shao, Y.; Kwok, L.-Y.; Bilige, M.; Mang, L.; Zhang, H. Angiotensin-converting enzyme inhibitory activity of Lactobacillus helveticus strains from traditional fermented dairy foods and antihypertensive effect of fermented milk of strain H9. J. Dairy Sci. 2014, 97, 6680–6692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Yu, Z.; Zhao, W.; Lin, S.; Wang, E.; Zhang, Y.; Hao, H.; Wang, Z.; Chen, F. Isolation and identification of angiotensin-converting enzyme inhibitory peptides from egg white protein hydrolysates. Food Chem. 2010, 122, 1159–1163. [Google Scholar] [CrossRef]
- Montone, C.M.; Chiozzi, R.Z.; Marchetti, N.; Cerrato, A.; Antonelli, M.; Capriotti, A.L.; Cavaliere, C.; Piovesana, S.; Laganà, A. Peptidomic approach for the identification of peptides with potential antioxidant and anti-hyperthensive effects derived from asparagus by-products. Molecules 2019, 24, 3627. [Google Scholar] [CrossRef] [Green Version]
- Shi, C.; Li, T.; Huang, J.; Khan, I.A.; Huang, M.; Zhou, G. Effect of processing conditions and simulated gastrointestinal digestion on the activity of angiotensin I-converting enzyme (ACE) inhibitory peptide derived from duck meat hydrolysate. Cyta-.J Food. 2019, 17, 393–399. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Chang, C.; Liu, H.; Li, B.; Yan, Q.; Jiang, Z. Identification of novel angiotensin I-converting enzyme (ACE) inhibitory peptides from wheat gluten hydrolysate by the protease of Pseudomonas aeruginosa. J. Funct. Foods 2020, 65, 103751. [Google Scholar] [CrossRef]
- Anekthanakul, K.; Senachak, J.; Hongsthong, A.; Charoonratana, T.; Ruengjitchatchawalya, M. Natural ACE inhibitory peptides discovery from Spirulina (Arthrospira platensis) strain C1. Peptides 2019, 118, 170107. [Google Scholar] [CrossRef]
- Brai, A.; Trivisani, C.I.; Vagaggini, C.; Stella, R.; Angeletti, R.; Iovenitti, G.; Francardi, V.; Dreassi, E. Proteins from Tenebrio molitor: An interesting functional ingredient and a source of ACE inhibitory peptides. Food Chem. 2022, 393, 133409. [Google Scholar] [CrossRef]
- Huang, Y.; Jia, F.; Zhao, J.; Hou, Y.; Hu, S.Q. Novel ACE Inhibitory Peptides Derived from Yeast Hydrolysates: Screening, Inhibition Mechanisms and Effects on HUVECs. J. Agric. Food Chem. 2021, 69, 2412–2421. [Google Scholar] [CrossRef]
- Li, T.; Shi, C.; Zhou, C.; Sun, X.; Ang, Y.; Dong, X.; Huang, M.; Zhou, G. Purification and characterization of novel antioxidant peptides from duck breast protein hydrolysates. LWT 2020, 125, 109215. [Google Scholar] [CrossRef]
- Cao, S.; Wang, Y.; Hao, Y.; Zhang, W.; Zhou, G. Antihypertensive effects in vitro and in vivo of novel angiotensin-converting enzyme inhibitory peptides from bovine bone gelatin hydrolysate. J. Agric. Food Chem. 2019, 68, 759–768. [Google Scholar] [CrossRef]
- Kong, X.; Zhang, L.; Song, W.; Zhang, C.; Hua, Y.; Chen, Y.; Li, X. Separation, identification and molecular binding mechanism of dipeptidyl peptidase IV inhibitory peptides derived from walnut (Juglans regia L.) protein. Food Chem. 2021, 347, 129062. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Ledesma, B.; Del Mar Contreras, M.; Recio, I. Antihypertensive peptides: Production, bioavailability and incorporation into foods. Adv. Colloid Interface Sci. 2011, 165, 23–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, J.; Chen, J.; Pan, B.S. ACE-inhibitory peptides identified from the muscle protein hydrolysate of hard clam (Meretrix lusoria). Process Biochem. 2008, 43, 743–747. [Google Scholar] [CrossRef]
- Wen, C.; Zhang, J.; Zhang, H.; Duan, Y.; Ma, H. Plant protein-derived antioxidant peptides: Isolation, identification, mechanism of action and application in food systems: A review. Trends Food Sci. Technol. 2020, 105, 308–322. [Google Scholar] [CrossRef]
- Okoye, C.O.; Ezeorba, T.P.; Okeke, E.S.; Okagu, I.U. Recent Findings on the Isolation, Identification and Quantification of Bioactive Peptides. Appl. Food Res. 2022, 2, 100065. [Google Scholar] [CrossRef]
- Natesh, R.; Schwager, S.L.; Sturrock, E.D.; Acharya, K.R. Crystal structure of the human angiotensin-converting enzyme–lisinopril complex. Nature 2003, 421, 551–554. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Aluko, R.E.; Nakai, S. Structural requirements of angiotensin I-converting enzyme inhibitory peptides: Quantitative structure-activity relationship modeling of peptides containing 4-10 amino acid residues. QSAR Comb. Sci. 2006, 25, 873–880. [Google Scholar] [CrossRef]
- Ondetti, M.A.; Rubin, B.; Cushman, D.W. Design of specific inhibitors of angiotensin-converting enzyme: New class of orally active antihypertensive agents. Science 1977, 196, 441–444. [Google Scholar] [CrossRef]
- Fan, H.; Liao, W.; Wu, J. Molecular interactions, bioavailability, and cellular mechanisms of angiotensin-converting enzyme inhibitory peptides. J. Food Biochem. 2019, 43, e12572. [Google Scholar] [CrossRef] [Green Version]
- Ghassem, M.; Arihara, K.; Babji, A.S.; Said, M.; Ibrahim, S. Purification and identification of ACE inhibitory peptides from Haruan (Channa striatus) myofibrillar protein hydrolysate using HPLC–ESI-TOF MS/MS. Food Chem. 2011, 129, 1770–1777. [Google Scholar] [CrossRef]
- Mirzapour, M.; Rezaei, K.; Sentandreu, M.A. Identification of potent ACE inhibitory peptides from wild almond proteins. J. Food Sci. 2017, 82, 2421–2431. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Qin, S.; Li, W. Purification and characterization of a novel angiotensin I-converting enzyme-inhibitory peptide derived from Alaska pollack skins. J. Food Sci. 2021, 86, 2457–2467. [Google Scholar] [CrossRef] [PubMed]
- You, H.; Wu, T.; Wang, W.; Li, Y.; Liu, X.; Ding, L. Preparation and identification of dipeptidyl peptidase IV inhibitory peptides from quinoa protein. Food Res. Int. 2022, 156, 111176. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, M.; Mirdamadi, S.; Ehsani, M.R.; Aminlari, M. Production of antioxidant and ACE-inhibitory peptides from Kluyveromyces marxianus protein hydrolysates: Purification and molecular docking. J. Food Drug. Anal. 2018, 26, 696–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baba, W.N.; Baby, B.; Mudgil, P.; Gan, C.; Vijayan, R.; Maqsood, S. Pepsin generated camel whey protein hydrolysates with potential antihypertensive properties: Identification and molecular docking of antihypertensive peptides. LWT 2021, 143, 111135. [Google Scholar] [CrossRef]
- Wei, G.; Zhao, Q.; Wang, D.; Fan, Y.; Shi, Y.; Huang, A. Novel ACE inhibitory, antioxidant and α-glucosidase inhibitory peptides identified from fermented rubing cheese through peptidomic and molecular docking. LWT 2022, 159, 113196. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, X.; Zhuang, Y.; Li, Y.; Shi, P.; Tian, H.; Li, X.; Chen, X. Isolation of novel ACE-inhibitory peptide from naked oat globulin hydrolysates in silico approach: Molecular docking, in vivo antihypertension and effects on renin and intracellular endothelin-1. J. Food Sci. 2020, 85, 1328–1337. [Google Scholar] [CrossRef]
- Cheung, H.; Wang, F.; Ondetti, M.; Sabo, E.; Cushman, D. Binding of peptide substrates and inhibitors of angiotensin-converting enzyme: Importance of the COOH-terminal dipeptide sequence. J. Biol. Chem. 1980, 255, 401–407. [Google Scholar] [CrossRef]
- Miyoshi, S.; Kaneko, T.; Ishikawa, H.; Tanaka, H.; Maruyama, S. Production of bioactive peptides from corn endosperm proteins by some proteases. Ann. N. Y. Acad. Sci. 1995, 750, 429–431. [Google Scholar] [CrossRef]
- Udenigwe, C.C.; Li, H.; Aluko, R.E. Quantitative structure–activity relationship modeling of renin-inhibiting dipeptides. Amino Acids 2012, 42, 1379–1386. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; Mooney, C.; Shields, D.C.; Fitzgerald, R.J. Inhibition of dipeptidyl peptidase IV and xanthine oxidase by amino acids and dipeptides. Food Chem. 2013, 141, 644–653. [Google Scholar] [CrossRef] [PubMed]




| No. | Sequence | Length | Mass/Da | Proteins | Charges | Score | Intensity | Hydrophobicity/% | Hydrophobic Amino Acid Location |
|---|---|---|---|---|---|---|---|---|---|
| 1 | VALSSLRP | 8 | 841.5022 | U3I3Y9 | 2 | 111.28 | 3.29’108 | 62.50 | 1,2,3,6,8 |
| 2 | QFQPGFSSS | 9 | 983.4349 | A0A7K7L595 | 2 | 113.7 | 1.37´108 | 33.33 | 2,4,6 |
| 3 | TTPSYVAFTDTER | 13 | 1486.694 | U3IT45 | 2 | 169.58 | 9.90´107 | 30.77 | 3,6,7,8 |
| 4 | STGEAFVQFASQEIAEK | 17 | 1840.884 | A0A493SXK6 | 2 | 136.99 | 4.10´107 | 41.18 | 5,6,7,9,10,14,15 |
| 5 | NPDDITNEEYGEFYK | 15 | 1832.774 | A0A7K7LEE6 | 2 | 114.72 | 1.05´107 | 13.33 | 5,13 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Z.; Guo, H.; Xu, Y.; Pius Bassey, A.; Ali, A.; Huang, M.; Huang, J. ACE Inhibitory Peptides Derived from Muscovy Duck (Cairina moschata) Plasma. Foods 2023, 12, 50. https://doi.org/10.3390/foods12010050
Zhu Z, Guo H, Xu Y, Pius Bassey A, Ali A, Huang M, Huang J. ACE Inhibitory Peptides Derived from Muscovy Duck (Cairina moschata) Plasma. Foods. 2023; 12(1):50. https://doi.org/10.3390/foods12010050
Chicago/Turabian StyleZhu, Zongshuai, Haoyu Guo, Yan Xu, Anthony Pius Bassey, Ahtisham Ali, Ming Huang, and Jichao Huang. 2023. "ACE Inhibitory Peptides Derived from Muscovy Duck (Cairina moschata) Plasma" Foods 12, no. 1: 50. https://doi.org/10.3390/foods12010050
APA StyleZhu, Z., Guo, H., Xu, Y., Pius Bassey, A., Ali, A., Huang, M., & Huang, J. (2023). ACE Inhibitory Peptides Derived from Muscovy Duck (Cairina moschata) Plasma. Foods, 12(1), 50. https://doi.org/10.3390/foods12010050