The Microbial Diversity of Non-Korean Kimchi as Revealed by Viable Counting and Metataxonomic Sequencing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Kimchi Production
2.2. pH Determination
2.3. Microbial Counts
2.4. RNA Extraction and cDNA Synthesis
2.5. Metataxonomic Sequencing and Bioinformatics Analysis
2.6. Data Analysis
3. Results
3.1. pH Determination
3.2. Microbiological Analyses
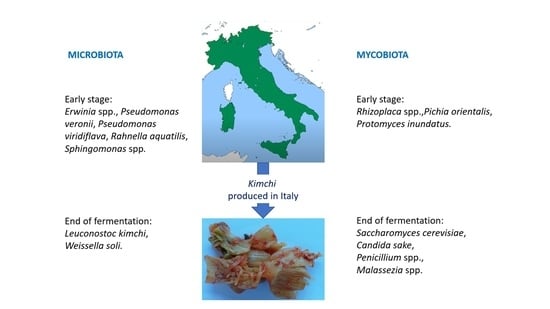
3.3. Microbiota Diversity
3.4. Mycobiota Diversity
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Misihairabgwi, J.; Cheikhyoussef, A. Traditional fermented foods and beverages of Namibia. J. Ethn. Foods 2017, 4, 145–153. [Google Scholar] [CrossRef]
- Ramos, C.L.; Schwan, R.F. Technological and nutritional aspects of indigenous Latin America fermented foods. Curr. Opin. Food Sci. 2017, 13, 97–102. [Google Scholar] [CrossRef]
- Majcherczyk, J.; Surówka, K. Effects of onion or caraway on the formation of biogenic amines during sauerkraut fermentation and refrigerated storage. Food Chem. 2019, 298, 125083. [Google Scholar] [CrossRef] [PubMed]
- Jaehae, L.; Kyeongsoon, H.; Chaelin, P.; Ilgwon, K.; Jeongwon, K.; Dukno, Y.; Montanari, M.; Naomichi, I.; Cwiertka, K.J.; Younghee, S.; et al. Humanistic Understanding of Kimchi and Kimjang Culture, 1st ed.; World Institute of Kimchi: Gwangju, Korea, 2015. [Google Scholar]
- Patra, J.K.; Das, G.; Paramithiotis, S.; Shin, H.S. Kimchi and other widely consumed traditional fermented foods of Korea: A review. Front. Microbiol. 2016, 7, 1493. [Google Scholar] [CrossRef] [PubMed]
- Jang, D.-J.; Chung, K.R.; Yang, H.J.; Kim, K.-S.; Kwon, D.Y. Discussion on the origin of kimchi, representative of Korean unique fermented vegetables. J. Ethn. Foods 2015, 2, 126–136. [Google Scholar] [CrossRef] [Green Version]
- Jung, J.Y.; Lee, S.H.; Jeon, C.O. Kimchi microflora: History, current status, and perspectives for industrial kimchi production. App. Microbiol. Biotechnol. 2014, 98, 2385–2393. [Google Scholar] [CrossRef]
- Lee, H.-W.; Yoon, S.-R.; Kim, S.-J.; Lee, H.M.; Lee, J.Y.; Lee, J.-H.; Kim, S.H.; Ha, J.-H. Identification of microbial communities, with a focus on foodborne pathogens, during kimchi manufacturing process using culture-independent and -dependent analyses. LWT Food Sci. Technol. 2017, 81, 153–159. [Google Scholar] [CrossRef]
- Chung, H.-K.; Shin, D.; Chung, K.R.; Choi, S.Y.; Woo, N. Recovering the royal cuisine in Chosun Dynasty and its esthetics. J. Ethn. Foods 2017, 4, 242–253. [Google Scholar] [CrossRef]
- Lee, M.; Song, J.H.; Jung, M.Y.; Lee, S.H.; Chang, J.Y. Large-scale targeted metagenomics analysis of bacterial ecological changes in 88 kimchi samples during fermentation. Food Microbiol. 2017b, 66, 173–183. [Google Scholar] [CrossRef]
- Cheigh, H. Production, characteristics and health functions of kimchi. Acta Hortic. 1999, 483, 405–420. [Google Scholar] [CrossRef]
- Jung, J.Y.; Lee, S.H.; Kim, J.M.; Park, M.S.; Bae, J.-W.; Hahn, Y.; Madsen, E.L.; Jeon, C.O. Metagenomic analysis of kimchi, a traditional Korean fermented food. App. Environ. Microbiol. 2011, 77, 2264–2274. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.-E.; Jang, J.-Y.; Lee, J.-H.; Park, H.-W.; Choi, H.-J.; Kim, T.-W. Starter cultures for kimchi fermentation. J. Microbiol. Biotechnol. 2015, 25, 559–568. [Google Scholar] [CrossRef]
- Torres, S.; Verón, H.; Contreras, L.; Isla, M.I. An overview of plant-autochthonous microorganisms and fermented vegetable foods. Food Sci. Human Wellness 2020, 9, 112–123. [Google Scholar] [CrossRef]
- Chang, J.-H.; Shim, Y.Y.; Cha, S.-K.; Chee, K.M. Probiotic characteristics of lactic acid bacteria isolated from kimchi. J. Appl. Microbiol. 2010, 109, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.-K.; Han, K.J.; Son, S.-H.; Eom, S.J.; Lee, S.-K.; Paik, H.-D. Multifunctional effect of probiotic Lactococcus lactis KC24 isolated from kimchi. LWT Food Sci. Technol. 2015b, 64, 1036–1041. [Google Scholar] [CrossRef]
- Lee, K.-H.; Bong, Y.-J.; Lee, H.A.; Kim, H.-Y.; Park, K.-Y. Probiotic effects of Lactobacillus plantarum and Leuconostoc mesenteroides isolated from kimchi. J. Korean Soc. Food Sci. Nutr. 2016, 45, 12–19. [Google Scholar] [CrossRef]
- Kim, M.-J.; Lee, H.-W.; Kim, J.Y.; Kang, S.E.; Roh, S.W.; Hong, S.W.; Yoo, S.R.; Kim, T.-W. Impact of fermentation conditions on the diversity of white colony-forming yeast and analysis of metabolite changes by white colony-forming yeast in kimchi. Food Res. Int. 2020, 136, 109315. [Google Scholar] [CrossRef] [PubMed]
- Belleggia, L.; Milanović, V.; Ferrocino, I.; Cocolin, L.; Naceur Haouet, M.; Scuota, S.; Maoloni, A.; Garofalo, C.; Cardinali, F.; Aquilanti, L.; et al. Is there any still undisclosed biodiversity in Ciauscolo salami? A new glance into the microbiota of an artisan production as revealed by high-throughput sequencing. Meat Sci. 2020, 165, 108128. [Google Scholar] [CrossRef] [Green Version]
- Haouet, M.N.; Altissimi, M.S.; Mercuri, M.L.; Baldassarri, C.; Osimani, A.; Clementi, F.; Ortenzi, R. Evaluation of the safety of Milano-type dry fermented sausages produced by a fast drying technology. Ital. J. Food Sci. 2017, 29, 1–9. [Google Scholar]
- Garofalo, C.; Bancalari, E.; Milanović, V.; Cardinali, F.; Osimani, A.; Sardaro, M.L.S.; Bottari, B.; Bernini, V.; Aquilanti, L.; Clementi, F.; et al. Study of the bacterial diversity of foods: PCR-DGGE versus LH-PCR. Int. J. Food Microbiol. 2017, 242, 24–36. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Mota-Gutierrez, J.; Ferrocino, I.; Rantsiou, K.; Cocolin, L. Metataxonomic comparison between internal transcribed spacer and 26S ribosomal large subunit (LSU) rDNA gene. Int. J. Food Microbiol. 2019, 290, 132–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of highthroughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, H.C. Healthy and safe Korean traditional fermented foods: Kimchi and chongkukjang. J. Ethn. Foods 2018, 5, 161–166. [Google Scholar] [CrossRef]
- Jeong, S.H.; Jung, J.Y.; Lee, S.H.; Jin, H.M.; Jeon, C.O. Microbial succession and metabolite changes during fermentation of dongchimi, traditional Korean watery kimchi. Int. J. Food Microbiol. 2013, 164, 46–53. [Google Scholar] [CrossRef]
- Song, H.S.; Whon, T.W.; Kim, J.; Lee, S.H.; Kim, J.Y.; Kim, Y.B.; Choi, H.-J.; Rhee, J.-K.; Roh, S.W. Microbial niches in raw ingredients determine microbial community assembly during kimchi fermentation. Food Chem. 2020, 318, 126481. [Google Scholar] [CrossRef]
- Hong, Y.; Yang, H.-S.; Chang, H.-C.; Kim, H.-Y. Comparison of bacterial community changes in fermenting kimchi at two different temperatures using a denaturing gradient gel electrophoresis analysis. J. Microbiol. Biotechnol. 2013, 23, 76–84. [Google Scholar] [CrossRef] [Green Version]
- Yamani, M.I. Fermentation of brined tunrnip roots using Lactobacillus plantarum and Leuconostoc mesenteroides starter cultures. World J. Microbiol. Biotechnol. 1993, 9, 176–179. [Google Scholar] [CrossRef]
- Hurtado, A.; Reguant, C.; Esteve-Zarzoso, B.; Bordons, A.; Rozès, N. Microbial population dynamics during the processing of Arbequina table olives. Food Res. Int. 2008, 41, 738–744. [Google Scholar] [CrossRef]
- Tamang, B.; Tamang, J.P. In situ fermentation dynamics during production of gundruk and khalpi, ethnic fermented vegetables products of the Himalayas. Indian J. Microbiol. 2010, 50, 93–98. [Google Scholar] [CrossRef] [Green Version]
- Wouters, D.; Bernaert, N.; Conjaerts, W.; Van Droogenbroeck, B.; De Loose, M.; De Vuyst, L. Species diversity, community dynamics, and metabolite kinetics of spontaneous leek fermentations. Food Microbiol. 2013, 33, 185–196. [Google Scholar] [CrossRef]
- Osimani, A.; Milanović, V.; Cardinali, F.; Garofalo, C.; Clementi, F.; Pasquini, M.; Riolo, P.; Ruschioni, S.; Isidoro, N.; Loreto, N.; et al. The bacterial biota of laboratory-reared edible mealworms (Tenebrio molitor L.): From feed to frass. Int. J. Food Microbiol. 2018, 272, 49–60. [Google Scholar] [CrossRef]
- Park, E.-J.; Chun, J.; Cha, C.-J.; Park, W.-S.; Jeon, C.O.; Bae, J.-W. Bacterial community analysis during fermentation of ten representative kinds of kimchi with barcoded pyrosequencing. Food Microbiol. 2012, 30, 197–204. [Google Scholar] [CrossRef]
- Wouters, D.; Grosu-Tudor, S.; Zamfir, M.; De Vuyst, L. Bacterial community dynamics, lactic acid bacteria species divesity and metabolite kinetics of traditional Romanian vegetable fermentations. J. Sci. Food Agric. 2013, 93, 749–760. [Google Scholar] [CrossRef]
- Chang, H.-W.; Kim, K.-H.; Nam, Y.-D.; Roh, S.W.; Kim, M.-S.; Jeon, C.O.; Oh, H.-M.; Bae, J.-W. Analysis of yeast and archaeal population dynamics in kimchi using denaturing gradient gel electrophoresis. Int. J. Food Microbiol. 2008, 126, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Elomari, M.; Coroler, L.; Hoste, B.; Gillis, M.; Izard, D.; Leclerc, H. DNA relatedness among Pseudomonas strains isolated from natural mineral waters and proposal of Pseudomonas veronii sp. nov. Int. J. Syst. Evol. Microbiol. 1996, 46, 1138–1144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, G.; Ottesen, A.; Bolten, S.; Wang, L.; Luo, Y.; Rideout, S.; Lyu, S.; Nou, X. Impact of routine sanitation on the microbiomes in a fresh produce processing facility. Int. J. Food Microbiol. 2019, 294, 31–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kahala, M.; Blasco, L.; Joutsjoki, V. Molecular characterization of spoilage bacteria as a means to observe the microbiological quality of carrot. J. Food Prot. 2012, 75, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Calvo, J.; Calvente, V.; de Orellano, M.E.; Benuzzi, D.; Sanz de Tosetti, M.I. Biological control of postharvest spoilage caused by Penicillium expansum and Botrytis cinerea in apple by using the bacterium Rahnella aquatilis. Int. J. Food Microbiol. 2007, 113, 251–257. [Google Scholar] [CrossRef]
- Lee, M.; Song, J.H.; Park, J.M.; Chang, J.Y. Bacterial diversity in Korean temple kimchi fermentation. Food Res. Int. 2019, 126, 108592. [Google Scholar] [CrossRef]
- Ragaert, P.; Devlieghere, F.; Debevere, J. Role of microbiological and physiological spoilage mechanisms during storage of minimally processed vegetables. Postharvest Biol. Technol. 2017, 44, 185–194. [Google Scholar] [CrossRef]
- Park, S.; Ji, Y.; Park, H.; Lee, K.; Park, H.; Beck, B.R.; Shin, H.; Holzapfel, W.H. Evaluation of functional properties of lactobacilli isolated from Korean white kimchi. Food Control. 2016, 69, 5–12. [Google Scholar] [CrossRef]
- Menon, R.R.; Kumari, S.; Kumar, P.; Verma, A.; Krishnamurthi, S.; Rameshkumar, N. Sphingomonas pokkalii sp. nov., a novel plant associated rhizobacterium isolated from a saline tolerant pokkali rice and its draft genome analysis. Syst. App. Microbiol. 2019, 42, 334–342. [Google Scholar] [CrossRef]
- Jung, M.Y.; Kim, T.-W.; Lee, C.; Kim, J.Y.; Song, H.S.; Kim, Y.B.; Ahn, S.W.; Kim, J.S.; Roh, S.W.; Lee, S.H. Role of jeotgal, a Korean traditional fermented fish sauce, in microbial dynamics and metabolite profiles during kimchi fermentation. Food Chem. 2018, 265, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Magnusson, J.; Jonsson, H.; Schnürer, J.; Roos, S. Weissella soli sp. nov., a lactic acid bacterium isolated from soil. Int. J. Syst. Evol. Microbiol. 2002, 52, 831–834. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Cho, Y.; Lee, Y.; Han, S.-K.; Kim, C.-G.; Choo, D.-W.; Kim, Y.-R.; Kim, H.-Y. A proteomic approach for rapid identification of Weissella species isolated from Korean fermented foods on MALDI-TOF MS supplemented with an in-house database. Int. J. Food Microbiol. 2017, 243, 9–15. [Google Scholar] [CrossRef]
- Yu, H.-S.; Jang, H.J.; Lee, N.-K.; Paik, H.-D. Evaluation of the probiotic characteristics and prophylactic potential of Weissella cibaria strains isolated from kimchi. LWT 2019, 112, 108229. [Google Scholar] [CrossRef]
- Oh, H.-M.; Cho, Y.-J.; Kim, B.K.; Roe, J.-H.; Kang, S.-O.; Nahm, B.H.; Jeong, G.; Han, H.-U.; Chun, J. Complete genome sequence analysis of Leuconostoc kimchii IMSNU 11154. J. Bacteriol. 2010, 192, 3844–3845. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Jung, J.Y.; Lee, S.H.; Jeon, C.O. Complete genome sequence of Leuconostoc kimchii strain C2, isolated from Kimchi. J. Bacteriol. 2011, 193, 5548. [Google Scholar] [CrossRef] [Green Version]
- Jung, J.Y.; Jeong, J.-W.; Lee, S.-Y.; Jin, H.M.; Choi, H.W.; Ryu, B.-G.; Han, S.-S.; Kang, H.K.; Chung, E.J.; Choi, K.-M. Complete genome sequence of Leuconostoc kimchii strain NKJ218, isolated from homemade kimchi. Microbiol. Resour. Announc. 2019, 8, e00367-19. [Google Scholar] [CrossRef] [Green Version]
- Choi, B.-R.; Kwon, E.-Y.; Kim, H.-J.; Choi, M.-S. Role of synbiotics containing D-allulose in the alteration of body fat and hepatic lipids in diet-induced obese mice. Nutrients 2018, 10, 1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jo, S.Y.; Choi, E.A.; Lee, J.J.; Chang, H.C. Characterization of starter kimchi fermented with Leuconostoc kimchii GJ2 and its cholesterol-lowering effects in rats fed a high-fat and high-cholesterol diet. J. Sci. Food Agric. 2015, 95, 2750–2756. [Google Scholar] [CrossRef] [PubMed]
- Schleifer, K.H. Leuconostocaceae fam. nov. Bergey’s Man. Syst. Archaea Bact. 2019. [Google Scholar] [CrossRef]
- Torres-Rodríguez, I.; Rodríguez-Alegría, M.E.; Miranda-Molina, A.; Giles-Gómez, M.; Conca Morales, R.; López-Munguía, A.; Bolívar, F.; Escalante, A. Screening and characterization of extracellular polysaccharides produced by Leuconostoc kimchii isolated from traditional fermented pulque beverage. Springerplus 2014, 3, 583. [Google Scholar] [CrossRef] [Green Version]
- Rizzello, C.G.; Coda, R.; Wang, Y.; Verni, M.; Kajala, I.; Katina, K.; Laitila, A. Characterization of indigenous Pediococcus pentosaceus, Leuconostoc kimchii, Weissella cibaria and Weissella confusa for faba bean bioprocessing. Int. J. Food Microbiol. 2019, 302, 24–34. [Google Scholar] [CrossRef]
- Leavitt, S.D.; Fankhauser, J.D.; Leavitt, D.H.; Porter, L.D.; Johnson, L.A.; St. Clair, L.L. Complex patterns of speciation in cosmopolitan “rock posy” lichens–Discovering and delimiting cryptic fungal species in the lichen-forming Rhizoplaca melanophthalma species-complex (Lecanoraceae, Ascomycota). Mol. Phylogenetics Evol. 2011, 59, 587–602. [Google Scholar] [CrossRef]
- Choi, M.H.; Park, Y.H. Growth of Pichia guilliermondii A9, an osmotolerant yeast, in waste brine generated from kimchi production. Bioresour. Technol. 1999, 70, 231–236. [Google Scholar] [CrossRef]








| Batch | Sampling Time (Days) | Mesophilic Aerobic Bacteria | Halophilic Mesophilic Aerobic Bacteria | Presumptive Lactobacilli | Presumptive Halophilic Lactobacilli | Presumptive Lactococci | Presumptive Halophilic Lactococci | Enterobacteriaceae | Yeasts | Halophilic Yeasts | Pseudomonadaceae |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | t0 | 5.2 ± 0.1 a | 5.3 ± 0.1 a | 3.5 ± 0.1 b | 3.5 ± 0.1 b | 5.0 ± 0.1 a | 4.9 ± 0.0 a | 1.7 ± 0.3 b | 1.3 ± 0.0 a | <1.0 b | 4.5 ± 0.2 a |
| t2 | 5.3 ± 0.0 a | 5.3 ± 0.1 a | 3.6 ± 0.5 b | 3.5 ± 0.5 b | 5.0 ± 0.1 a | 5.1 ± 0.1 a | 2.3 ± 0.6 b | 1.4 ± 0.1 a | 1.4 ± 0.1 a | 5.2 ± 0.1 a | |
| t5 | 5.2 ± 0.0 a | 5.2 ± 0.0 a | 4.2 ± 0.0 b | 4.2 ± 0.0 b | 5.0 ± 0.1 a | 5.1 ± 0.2 a | 3.5 ± 0.5 a | 1.1 ± 0.1 a | 1.2 ± 0.3 a | 4.8 ± 0.3 a | |
| t15 | 5.3 ± 0.1 a | 5.3 ± 0.0 a | 4.8 ± 0.2 b | 4.8 ± 0.1 b | 5.2 ± 0.0 a | 4.8 ± 0.1 a | 3.4 ± 0.2 a | <1.0 b | <1.0 b | 5.1 ± 0.1 a | |
| t36 | 6.3 ± 0.1 a | 5.9 ± 0.2 a | 6.3 ± 0.0 a | 6.2 ± 0.2 a | 5.0 ± 0.2 a | 4.9 ± 0.2 a | <1.0 c | <1.0 b | <1.0 b | <1.0 b | |
| t43 | 5.6 ± 0.0 a | 5.3 ± 0.0 a | 6.3 ± 0.1 a | 6.4 ± 0.1 a | 5.0 ± 0.2 a | 5.1 ± 0.1 a | <1.0 c | <1.0 b | <1.0 b | <1.0 b | |
| t50 | 5.5 ± 0.0 a | 5.4 ± 0.0 a | 6.2 ± 0.2 a | 5.6 ± 0.1 a | 4.9 ± 0.3 a | 5.0 ± 0.1 a | <1.0 c | <1.0 b | <1.0 b | <1.0 b | |
| t57 | 5.4 ± 0.0 a | 5.3 ± 0.0 a | 4.6 ± 0.1 b | 4.7 ± 0.1 b | 5.0 ± 0.2 a | 5.0 ± 0.1 a | <1.0 c | <1.0 b | <1.0 b | <1.0 b | |
| 2 | t0 | 5.7 ± 0.3 a | 5.6 ± 0.2 a | 2.1 ± 0.2 c | 2.4 ± 0.0 b | 5.5 ± 0.3 a | 5.4 ± 0.1 a | 2.8 ± 0.2 a | 2.5 ± 0.1 a | 1.9 ± 0.0 a | 4.9 ± 0.1 a |
| t2 | 5.3 ± 0.1 a | 5.3 ± 0.0 a | 1.5 ± 0.1 c | 1.4 ± 0.2 b | 5.2 ± 0.1 a | 5.1 ± 0.2 a | 2.0 ± 0.2 a | 2.1 ± 0.0 a | 1.7 ± 0.0 a | 4.2 ± 0.0 a | |
| t5 | 5.2 ± 0.1 a | 5.2 ± 0.1 a | 1.2 ± 0.0 c | 1.2 ± 0.1 b | 5.0 ± 0.1 a | 4.8 ± 0.1 a | 1.8 ± 0.1 a | 1.5 ± 0.2 b | 1.0 ± 0.2 b | 4.9 ± 0.0 a | |
| t15 | 5.1 ± 0.1 a | 4.8 ± 0.1 a | 2.0 ± 0.1 c | 1.5 ± 0.1 b | 4.9 ± 0.0 a | 4.4 ± 0.0 a | 2.4 ± 0.3 a | 1.0 ± 0.0 b | < 1.0 b | 4.9 ± 0.0 a | |
| t36 | 5.1 ± 0.1 a | 5.1 ± 0.2 a | 4.8 ± 0.7 b | 2.5 ± 0.8 b | 4.8 ± 0.0 a | 4.3 ± 0.4 a | 1.6 ± 0.6 a | <1.0 c | <1.0 b | <1.0 b | |
| t43 | 4.9 ± 0.0 a | 4.8 ± 0.2 a | 7.3 ± 0.2 a | 4.2 ± 0.3 a | 4.5 ± 0.3 a | 4.4 ± 0.3 a | <1.0 c | <1.0 c | <1.0 b | <1.0 b | |
| t50 | 4.9 ± 0.0 a | 4.7 ± 0.0 a | 7.3 ± 0.0 a | 5.0 ± 0.4 a | 4.6 ± 0.1 a | 4.6 ± 0.0 a | <1.0 c | <1.0 c | <1.0 b | <1.0 b | |
| t57 | 4.9 ± 0.1 a | 4.9 ± 0.1 a | 6.4 ± 0.5 a | 4.3 ± 0.1 a | 4.2 ± 0.1 a | 4.2 ± 0.1 a | <1.0 c | <1.0 c | <1.0 b | <1.0 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maoloni, A.; Ferrocino, I.; Milanović, V.; Cocolin, L.; Corvaglia, M.R.; Ottaviani, D.; Bartolini, C.; Talevi, G.; Belleggia, L.; Cardinali, F.; et al. The Microbial Diversity of Non-Korean Kimchi as Revealed by Viable Counting and Metataxonomic Sequencing. Foods 2020, 9, 1568. https://doi.org/10.3390/foods9111568
Maoloni A, Ferrocino I, Milanović V, Cocolin L, Corvaglia MR, Ottaviani D, Bartolini C, Talevi G, Belleggia L, Cardinali F, et al. The Microbial Diversity of Non-Korean Kimchi as Revealed by Viable Counting and Metataxonomic Sequencing. Foods. 2020; 9(11):1568. https://doi.org/10.3390/foods9111568
Chicago/Turabian StyleMaoloni, Antonietta, Ilario Ferrocino, Vesna Milanović, Luca Cocolin, Maria Rita Corvaglia, Donatella Ottaviani, Chiara Bartolini, Giulia Talevi, Luca Belleggia, Federica Cardinali, and et al. 2020. "The Microbial Diversity of Non-Korean Kimchi as Revealed by Viable Counting and Metataxonomic Sequencing" Foods 9, no. 11: 1568. https://doi.org/10.3390/foods9111568
APA StyleMaoloni, A., Ferrocino, I., Milanović, V., Cocolin, L., Corvaglia, M. R., Ottaviani, D., Bartolini, C., Talevi, G., Belleggia, L., Cardinali, F., Marabini, R., Aquilanti, L., & Osimani, A. (2020). The Microbial Diversity of Non-Korean Kimchi as Revealed by Viable Counting and Metataxonomic Sequencing. Foods, 9(11), 1568. https://doi.org/10.3390/foods9111568