A Decision Support System for Diagnosis of COVID-19 from Non-COVID-19 Influenza-like Illness Using Explainable Artificial Intelligence
Abstract
:1. Introduction
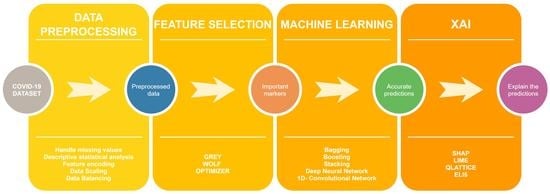
- We have collected our own COVID-19 dataset containing patient data of COVID-19 and non-COVID-19 influenza-like illness (ILI) patients from two Manipal hospitals in India. Prior ethical clearance has also been obtained to conduct this study.
- The statistical tool “JAMOVI” has been used to conduct a descriptive statistical analysis of the data.
- The grey wolf optimizer has been utilized for feature selection to choose the most essential clinical markers.
- Different ML algorithms have been tested to predict COVID-19 diagnosis. The algorithms have been further stacked on multiple levels to improve accuracy. Deep learning models such as deep learning networks (DNN) and one-dimensional convolutional neural networks (1D-CNN) have also been utilized to test model effectiveness.
- XAI techniques such as SHAP, LIME, Eli5 and QLattice have made the models more understandable and interpretable.
- Further discussion about COVID-19 diagnosis using important clinical markers is presented.
2. Materials and Methods
2.1. Dataset Description
2.2. Dataset Preprocessing
2.3. Grey Wolf Optimizer for Feature Selection
2.4. Machine Learning Terminologies and Pipeline
3. Results and Discussion
3.1. Performance Measures
3.2. Model Evaluation Using Machine Learning and Deep Learning
3.3. Explainable Artificial Intelligence (XAI) to Interpret Results
3.4. Further Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Betthäuser, B.A.; Bach-Mortensen, A.M.; Engzell, P. A systematic review and meta-analysis of the evidence on learning during the COVID-19 pandemic. Nat. Hum. Behav. 2023, 7, 375–385. [Google Scholar] [CrossRef]
- WHO. Coronavirus Disease (COVID-19). 2021. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 27 January 2023).
- Ciotti, M.; Ciccozzi, M.; Terrinoni, A.; Jiang, W.C.; Wang, C.B.; Bernardini, S. The COVID-19 pandemic. Crit. Rev. Clin. Lab. Sci. 2020, 57, 365–388. [Google Scholar] [CrossRef] [PubMed]
- Ndwandwe, D.; Wiysonge, C.S. COVID-19 vaccines. Curr. Opin. Immunol. 2021, 71, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Alimohamadi, Y.; Sepandi, M.; Taghdir, M.; Hosamirudsari, H. Determine the most common clinical symptoms in COVID-19 patients: A systematic review and meta-analysis. J. Prev. Med. Hyg. 2020, 61, E304. [Google Scholar] [CrossRef]
- Lesbon, J.C.; Poleti, M.D.; de Mattos Oliveira, E.C.; Patané, J.S.; Clemente, L.G.; Viala, V.L.; Ribeiro, G.; Giovanetti, M.; de Alcantara, L.C.; Teixeira, O.; et al. Correction: Lesbon et al. Nucleocapsid (N) Gene Mutations of SARS-CoV-2 Can Affect Real-Time RT-PCR Diagnostic and Impact False-Negative Results. Viruses 2021, 13, 2474. [Google Scholar] [CrossRef] [PubMed]
- Alves, M.A.; Castro, G.Z.; Oliveira, B.A.; Ferreira, L.A.; Ramírez, J.A.; Silva, R.; Guimarães, F.G. Explaining machine learning based diagnosis of COVID-19 from routine blood tests with decision trees and criteria graphs. Comput. Biol. Med. 2021, 132, 104335. [Google Scholar] [CrossRef]
- Gopatoti, A.; Vijayalakshmi, P. CXGNet: A tri-phase chest X-ray image classification for COVID-19 diagnosis using deep CNN with enhanced grey-wolf optimizer. Biomed. Signal Process. Control. 2022, 77, 103860. [Google Scholar] [CrossRef]
- Lou, L.; Xia, W.; Sun, Z.; Quan, S.; Yin, S.; Gao, Z.; Lin, C. COVID-19 mortality prediction using ensemble learning and grey wolf optimization. PeerJ Comput. Sci. 2023, 9, e1209. [Google Scholar] [CrossRef]
- AlJame, M.; Ahmad, I.; Imtiaz, A.; Mohammed, A. Ensemble learning model for diagnosing COVID-19 from routine blood tests. Inform. Med. Unlocked 2020, 21, 100449. [Google Scholar] [CrossRef]
- Alballa, N.; Al-Turaiki, I. Machine learning approaches in COVID-19 diagnosis, mortality, and severity risk prediction: A review. Inform. Med. Unlocked 2021, 24, 100564. [Google Scholar] [CrossRef]
- Giotta, M.; Trerotoli, P.; Palmieri, V.O.; Passerini, F.; Portincasa, P.; Dargenio, I.; Mokhtari, J.; Montagna, M.T.; De Vito, D. Application of a Decision Tree Model to Predict the Outcome of Non-Intensive Inpatients Hospitalized for COVID-19. Int. J. Environ. Res. Public Health 2022, 19, 13016. [Google Scholar] [CrossRef]
- Rahman, T.; Khandakar, A.; Abir, F.F.; Faisal, M.A.; Hossain, M.S.; Podder, K.K.; Abbas, T.O.; Alam, M.F.; Kashem, S.B.; Islam, M.T.; et al. QCovSML: A reliable COVID-19 detection system using CBC biomarkers by a stacking machine learning model. Comput. Biol. Med. 2022, 143, 105284. [Google Scholar] [CrossRef]
- Fang, L.; Liang, X. ISW-LM: An intensive symptom weight learning mechanism for early COVID-19 diagnosis. Comput. Biol. Med. 2022, 146, 105615. [Google Scholar] [CrossRef]
- Rostami, M.; Oussalah, M. A novel explainable COVID-19 diagnosis method by integration of feature selection with random forest. Inform. Med. Unlocked 2022, 30, 100941. [Google Scholar] [CrossRef] [PubMed]
- Thimoteo, L.M.; Vellasco, M.M.; Amaral, J.; Figueiredo, K.; Yokoyama, C.L.; Marques, E. Explainable artificial intelligence for COVID-19 diagnosis through blood test variables. J. Control. Autom. Electr. Syst. 2022, 33, 625–644. [Google Scholar] [CrossRef]
- Loh, H.W.; Ooi, C.P.; Seoni, S.; Barua, P.D.; Molinari, F.; Acharya, U.R. Application of explainable artificial intelligence for healthcare: A systematic review of the last decade (2011–2022). Comput. Methods Programs Biomed. 2022, 226, 107161. [Google Scholar] [CrossRef] [PubMed]
- Jahmunah, V.; Ng, E.Y.; Tan, R.S.; Oh, S.L.; Acharya, U.R. Explainable detection of myocardial infarction using deep learning models with Grad-CAM technique on ECG signals. Comput. Biol. Med. 2022, 146, 105550. [Google Scholar] [CrossRef] [PubMed]
- Şahin, M.; Aybek, E. Jamovi: An easy to use statistical software for the social scientists. Int. J. Assess. Tools Educ. 2019, 6, 670–692. [Google Scholar] [CrossRef] [Green Version]
- Cao, X.H.; Stojkovic, I.; Obradovic, Z. A robust data scaling algorithm to improve classification accuracies in biomedical data. BMC Bioinform. 2016, 17, 359. [Google Scholar] [CrossRef] [Green Version]
- Al-Shehari, T.; Alsowail, R.A. An insider data leakage detection using one-hot encoding, synthetic minority oversampling and machine learning techniques. Entropy 2021, 23, 1258. [Google Scholar] [CrossRef]
- Han, H.; Wang, W.Y.; Mao, B.H. Borderline-SMOTE: A new over-sampling method in imbalanced data sets learning. In Proceedings of the Advances in Intelligent Computing: International Conference on Intelligent Computing, ICIC 2005, Hefei, China, 23–26 August 2005; Springer: Berlin/Heidelberg, Germany, 2005. Part I. pp. 878–887. [Google Scholar] [CrossRef]
- Remeseiro, B.; Bolon-Canedo, V. A review of feature selection methods in medical applications. Comput. Biol. Med. 2019, 112, 103375. [Google Scholar] [CrossRef] [PubMed]
- Al-Tashi, Q.; Md Rais, H.; Abdulkadir, S.J.; Mirjalili, S.; Alhussian, H. A review of grey wolf optimizer-based feature selection methods for classification. In Evolutionary Machine Learning Techniques: Algorithms and Applications; Springer: Singapore, 2020; pp. 273–286. [Google Scholar] [CrossRef]
- Mirjalili, S.; Mirjalili, S.M.; Lewis, A. Grey wolf optimizer. Adv. Eng. Softw. 2014, 69, 46–61. [Google Scholar] [CrossRef] [Green Version]
- Too, J. Github. Available online: https://github.com/JingweiToo/Wrapper-Feature-Selection-Toolbox-Python (accessed on 30 January 2023).
- Alom, M.Z.; Taha, T.M.; Yakopcic, C.; Westberg, S.; Sidike, P.; Nasrin, M.S.; Hasan, M.; Van Essen, B.C.; Awwal, A.A.; Asari, V.K. A state-of-the-art survey on deep learning theory and architectures. Electronics 2019, 8, 292. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, A.A.; Ali, W.; Abdullah, T.A.; Malebary, S.J. Classifying Cardiac Arrhythmia from ECG Signal Using 1D CNN Deep Learning Model. Mathematics 2023, 11, 562. [Google Scholar] [CrossRef]
- Baptista, M.L.; Goebel, K.; Henriques, E.M. Relation between prognostics predictor evaluation metrics and local interpretability SHAP values. Artif. Intell. 2022, 306, 103667. [Google Scholar] [CrossRef]
- Dieber, J.; Kirrane, S. Why model why? Assessing the strengths and limitations of LIME. arXiv 2020, arXiv:2012.00093. [Google Scholar] [CrossRef]
- Kuzlu, M.; Cali, U.; Sharma, V.; Güler, Ö. Gaining insight into solar photovoltaic power generation forecasting utilizing explainable artificial intelligence tools. IEEE Access 2020, 8, 187814–187823. [Google Scholar] [CrossRef]
- Bharadi, V. QLattice Environment and Feyn QGraph Models—A New Perspective Toward Deep Learning. In Emerging Technologies for Healthcare: Internet of Things and Deep Learning Models; Scrivener: Cornwall, UK, 2021; pp. 69–92. [Google Scholar] [CrossRef]
- Kumar-M, P.; Mishra, S.; Jha, D.K.; Shukla, J.; Choudhury, A.; Mohindra, R.; Mandavdhare, H.S.; Dutta, U.; Sharma, V. Coronavirus disease (COVID-19) and the liver: A comprehensive systematic review and meta-analysis. Hepatol. Int. 2020, 14, 711–722. [Google Scholar] [CrossRef]
- Chen, L.Z.; Lin, Z.H.; Chen, J.; Liu, S.S.; Shi, T.; Xin, Y.N. Can elevated concentrations of ALT and AST predict the risk of ‘recurrence’of COVID-19? Epidemiol. Infect. 2020, 148, e218. [Google Scholar] [CrossRef]
- Bari, A.; Ch, A.; Bano, I.; Saqlain, N. Is leukopenia and lymphopenia a characteristic feature of COVID-19 in children? Pak. J. Med. Sci. 2021, 37, 869. [Google Scholar] [CrossRef]
- Shah, M.; Kakar, A.; Gogia, A.; Langer, S. Convalescent plasma, cytomegalovirus infection, and persistent leukopenia in COVID-19 recovery phase: What is the link? J. Postgrad. Med. 2021, 67, 100. [Google Scholar] [CrossRef] [PubMed]
- Kheir, M.; Saleem, F.; Wang, C.; Mann, A.; Chua, J. Higher albumin levels on admission predict better prognosis in patients with confirmed COVID-19. PLoS ONE 2021, 16, e0248358. [Google Scholar] [CrossRef] [PubMed]
- Harte, J.V.; Mykytiv, V. A panhaemocytometric approach to COVID-19: A retrospective study on the importance of monocyte and neutrophil population data on Sysmex XN-series analysers. Clin. Chem. Lab. Med. (CCLM) 2021, 59, e169–e172. [Google Scholar] [CrossRef] [PubMed]
- Murdaca, G.; Di Gioacchino, M.; Greco, M.; Borro, M.; Paladin, F.; Petrarca, C.; Gangemi, S. Basophils and mast cells in COVID-19 pathogenesis. Cells 2021, 10, 2754. [Google Scholar] [CrossRef]
- Reusch, N.; De Domenico, E.; Bonaguro, L.; Schulte-Schrepping, J.; Baßler, K.; Schultze, J.L.; Aschenbrenner, A.C. Neutrophils in COVID-19. Front. Immunol. 2021, 12, 652470. [Google Scholar] [CrossRef]
- Huang, W.; Berube, J.; McNamara, M.; Saksena, S.; Hartman, M.; Arshad, T.; Bornheimer, S.J.; O′Gorman, M. Lymphocyte subset counts in COVID-19 patients: A meta-analysis. Cytom. Part A 2020, 97, 772–776. [Google Scholar] [CrossRef]
- Li, X.; Liu, C.; Mao, Z.; Xiao, M.; Wang, L.; Qi, S.; Zhou, F. Predictive values of neutrophil-to-lymphocyte ratio on disease severity and mortality in COVID-19 patients: A systematic review and meta-analysis. Crit. Care 2020, 24, 647. [Google Scholar] [CrossRef]
- Ok, F.; Erdogan, O.; Durmus, E.; Carkci, S.; Canik, A. Predictive values of blood urea nitrogen/creatinine ratio and other routine blood parameters on disease severity and survival of COVID-19 patients. J. Med. Virol. 2021, 93, 786–793. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, Y.; Zhao, X.; Wang, L.; Liu, F.; Wang, T.; Ye, D.; Lv, Y. Diagnostic performance of a blood urea nitrogen to creatinine ratio-based nomogram for predicting in-hospital mortality in COVID-19 patients. Risk Manag. Healthc. Policy 2021, 12, 117–128. [Google Scholar] [CrossRef]
- Roland, T.; Böck, C.; Tschoellitsch, T.; Maletzky, A.; Hochreiter, S.; Meier, J.; Klambauer, G. Domain shifts in machine learning based COVID-19 diagnosis from blood tests. J. Med. Syst. 2022, 46, 23. [Google Scholar] [CrossRef]
- Rikan, S.B.; Azar, A.S.; Ghafari, A.; Mohasefi, J.B.; Pirnejad, H. COVID-19 diagnosis from routine blood tests using artificial intelligence techniques. Biomed. Signal Process. Control. 2022, 72, 103263. [Google Scholar] [CrossRef] [PubMed]
- de Freitas Barbosa, V.A.; Gomes, J.C.; de Santana, M.A.; Albuquerque, J.E.; de Souza, R.G.; de Souza, R.E.; dos Santos, W.P. Heg. IA: An intelligent system to support diagnosis of COVID-19 based on blood tests. Res. Biomed. Eng. 2021, 38, 99–116. [Google Scholar] [CrossRef]
- Bartenschlager, C.C.; Ebel, S.S.; Kling, S.; Vehreschild, J.; Zabel, L.T.; Spinner, C.D.; Schuler, A.; Heller, A.R.; Borgmann, S.; Hoffmann, R.; et al. COVIDAL: A machine learning classifier for digital COVID-19 diagnosis in German hospitals. ACM Trans. Manag. Inf. Syst. 2022, 14, 1–16. [Google Scholar] [CrossRef]













| Sl. No | Marker | Attribute Datatype | Description | Sl. No | Marker | Attribute Datatype | Description |
|---|---|---|---|---|---|---|---|
| 1 | Age | Demographic/Continuous | Age of a patient (In years) | 13 | Creatinine | Clinical/Continuous | It is an amino acid commonly found in muscles and brain. Higher levels of creatinine indicate damage to the kidney (mg/dL). |
| 2 | Gender | Demographic/Categorical | Gender of the patient (Male/ Female) | 14 | Sodium | Clinical/Continuous | Electrolytes which help body function by maintaining blood and volume. Higher levels of sodium can lead to hypertension (mmol/L). |
| 3 | Hemoglobin (Hb) | Clinical/Continuous | It carries oxygen to the organs of the body. It is a part of red blood cells (RBC) (gram/dL). | 15 | Potassium | Clinical/Continuous | Electrolytes which help body function by maintaining blood and volume. Lower levels of potassium can lead to hypertension (mmol/L). |
| 4 | Hematocrit | Clinical/Continuous | It indicates the proportion of RBC in blood (in %). | 16 | Total Bilirubin | Clinical/Continuous | It is a combination of direct and indirect bilirubin (mg/dL). |
| 5 | Total White blood cells (TWBC) | Clinical/Continuous | It fights infection and is a part of the immune system (103/microliter). | 17 | Direct Bilirubin | Clinical/Continuous | RBC’s are broken down by the body, creating a chemical called bilirubin (mg/dL). |
| 6 | Neutrophil | Clinical/Continuous | A type of WBC. Higher levels of neutrophil indicate an infection (% -Differential count). | 18 | Aspartate transaminase (AST) | Clinical/Continuous | It is an enzyme present in the liver. Higher levels of AST indicate damage to the liver (IU/L). |
| 7 | Lymphocyte | Clinical/Continuous | A type of WBC. Lower levels of lymphocyte indicate an infection (% -Differential count). | 19 | Alanine transaminase (ALT) | Clinical/Continuous | It is an enzyme present in the liver. Higher levels of AST indicate damage to the liver (IU/L). |
| 8 | NLR (Neutrophil to Lymphocyte ratio) | Clinical/Continuous | Number of neutrophils per lymphocytes. Higher levels of NLR indicate an infection (Whole number). | 20 | Alkaline phosphatase (ALP) | Clinical/Continuous | It is an enzyme present in the liver. Higher levels of AST indicate damage to the liver (IU/L). |
| 9 | Monocyte | Clinical/Continuous | A type of WBC. Varying levels of monocyte indicate infection in the body. | 21 | Protein | Clinical/Continuous | Total protein present in our blood (g/dL). |
| 10 | Eosinophil | Clinical/Continuous | A type of WBC. Varying levels of monocyte indicate infection in the body. (% -Differential count) | 22 | Albumin | Clinical/Continuous | A protein present in blood. Lower levels can indicate damage to kidneys or liver (g/dL). |
| 11 | Hemoglobin A1c (HbA1c) | Clinical/Continuous | It reveals the median blood sugar over a period of two to three months. Higher levels of HbA1c indicate diabetes (In %). | 23 | Urea | Clinical/Continuous | It is a main component of urine and removes unnecessary nitrogen. Higher levels of urea indicate damage to the kidney (mg/dL). |
| 12 | Basophil | Clinical/Continuous | A type of WBC (% -Differential count). | 24 | RT-PCR test results | Clinical/Categorical | Results of the RT-PCR test (COVID-19 positive/COVID-19 negative) |
| Feature | Class Label | Mean | Median | SD | IQR | Range | Minimum | Maximum | 25th | 50th | 75th |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Age | ILI (COVID-19 negative) | 52.711 | 54 | 19.929 | 32.75 | 82 | 18 | 100 | 35.25 | 54 | 68 |
| COVID-19 positive | 55.108 | 58 | 17.8 | 25 | 81 | 18 | 99 | 43 | 58 | 68 | |
| Hb(Hemoglobin) | ILI (COVID-19 negative) | 12.305 | 12.4 | 1.824 | 1.675 | 11.2 | 6.1 | 17.3 | 11.6 | 12.4 | 13.275 |
| COVID-19 positive | 12.718 | 12.9 | 2.158 | 2.65 | 15 | 3.7 | 18.7 | 11.55 | 12.9 | 14.2 | |
| PCV%(Haemtocrit) | ILI (COVID-19 negative) | 36.406 | 36.5 | 5.133 | 4.675 | 31.75 | 19.45 | 51.2 | 34.2 | 36.5 | 38.875 |
| COVID-19 positive | 37.762 | 38 | 6.274 | 7.8 | 48.5 | 9 | 57.5 | 34.2 | 38 | 42 | |
| TWBC | ILI (COVID-19 negative) | 8.497 | 7.95 | 4.316 | 2.425 | 33.9 | 1.2 | 35.1 | 6.55 | 7.95 | 8.975 |
| COVID-19 positive | 8.449 | 6.5 | 5.995 | 5.1 | 58.8 | 0.2 | 59 | 4.9 | 6.5 | 10 | |
| Neutrophil | ILI (COVID-19 negative) | 66.599 | 67.2 | 12.854 | 14.525 | 65.6 | 28.1 | 93.7 | 60.05 | 67.2 | 74.575 |
| COVID-19 positive | 72.977 | 74 | 14.207 | 20.9 | 87.36 | 10.64 | 98 | 63.95 | 74 | 84.85 | |
| Lymphocyte | ILI (COVID-19 negative) | 22.342 | 21.8 | 11.159 | 13.625 | 53.5 | 2.5 | 56 | 15 | 21.8 | 28.625 |
| COVID-19 positive | 17.721 | 15.7 | 11.923 | 17 | 90 | 1 | 91 | 8 | 15.7 | 25 | |
| NLR | ILI (COVID-19 negative) | 4.615 | 3 | 5.4 | 2.75 | 40 | 1 | 41 | 2 | 3 | 4.75 |
| COVID-19 positive | 8.242 | 4 | 11.118 | 8 | 92 | 1 | 93 | 2 | 4 | 10 | |
| Monocyte | ILI (COVID-19 negative) | 8.223 | 7.8 | 3.218 | 3.075 | 26.6 | 1 | 27.6 | 6.625 | 7.8 | 9.7 |
| COVID-19 positive | 7.761 | 7.5 | 3.772 | 4.9 | 20.8 | 0.2 | 21 | 5.1 | 7.5 | 10 | |
| Eosinophil | ILI (COVID-19 negative) | 1.99 | 1.5 | 1.904 | 1.725 | 8.3 | 0 | 8.3 | 0.8 | 1.5 | 2.525 |
| COVID-19 positive | 0.698 | 0.2 | 1.355 | 0.6 | 13.9 | 0 | 13.9 | 0.1 | 0.2 | 0.7 | |
| Basophil | ILI (COVID-19 negative) | 0.492 | 0.4 | 0.423 | 0.3 | 2.5 | 0 | 2.5 | 0.3 | 0.4 | 0.6 |
| COVID-19 positive | 0.316 | 0.2 | 0.287 | 0.2 | 4 | 0 | 4 | 0.2 | 0.2 | 0.4 | |
| Urea | ILI (COVID-19 negative) | 26.839 | 21.5 | 22.294 | 5.75 | 232 | 8 | 240 | 19.25 | 21.5 | 25 |
| COVID-19 positive | 36.745 | 26 | 35.139 | 22 | 242.3 | 0.7 | 243 | 19 | 26 | 41 | |
| Creatinine | ILI (COVID-19 negative) | 0.938 | 0.8 | 0.681 | 0.2 | 7.3 | 0.4 | 7.7 | 0.7 | 0.8 | 0.9 |
| COVID-19 positive | 1.211 | 0.9 | 1.383 | 0.4 | 14.8 | 0.2 | 15 | 0.7 | 0.9 | 1.1 | |
| Sodium | ILI (COVID-19 negative) | 133.911 | 135 | 5.163 | 3.75 | 30 | 112 | 142 | 132.25 | 135 | 136 |
| COVID-19 positive | 135.526 | 136 | 5.531 | 7 | 56 | 111 | 167 | 132 | 136 | 139 | |
| Potassium | ILI (COVID-19 negative) | 4.126 | 4.1 | 0.387 | 0.3 | 2.8 | 3.2 | 6 | 4 | 4.1 | 4.3 |
| COVID-19 positive | 4.245 | 4.2 | 0.659 | 0.8 | 5.9 | 2.1 | 8 | 3.8 | 4.2 | 4.6 | |
| T. Bilirubin | ILI (COVID-19 negative) | 0.716 | 0.5 | 1.127 | 0 | 12.2 | 0.2 | 12.4 | 0.5 | 0.5 | 0.5 |
| COVID-19 positive | 0.695 | 0.5 | 1.129 | 0.38 | 21 | 0 | 21 | 0.32 | 0.5 | 0.7 | |
| D.Bilirubin | ILI (COVID-19 negative) | 0.362 | 0.2 | 0.899 | 0 | 9.6 | 0.1 | 9.7 | 0.2 | 0.2 | 0.2 |
| COVID-19 positive | 0.341 | 0.2 | 0.731 | 0.2 | 11.96 | 0.04 | 12 | 0.1 | 0.2 | 0.3 | |
| AST | ILI (COVID-19 negative) | 46.719 | 33 | 62.99 | 0 | 589 | 10 | 599 | 33 | 33 | 33 |
| COVID-19 positive | 55.941 | 39 | 65.605 | 36 | 900.8 | 0.2 | 901 | 26 | 39 | 62 | |
| ALT | ILI (COVID-19 negative) | 41.648 | 35 | 34.909 | 2.375 | 257 | 9 | 266 | 33.375 | 35 | 35.75 |
| COVID-19 positive | 46.095 | 32 | 58.912 | 30 | 696.5 | 3.5 | 700 | 20 | 32 | 50 | |
| ALP | ILI (COVID-19 negative) | 95.622 | 89 | 44.133 | 0 | 469 | 35 | 504 | 89 | 89 | 89 |
| COVID-19 positive | 95.135 | 81 | 62.826 | 39 | 880 | 5 | 885 | 65 | 81 | 104 | |
| Protein | ILI (COVID-19 negative) | 7.021 | 7 | 0.414 | 0 | 3.1 | 5.9 | 9 | 7 | 7 | 7 |
| COVID-19 positive | 6.893 | 7 | 0.685 | 0.6 | 9.2 | 3.2 | 12.4 | 6.6 | 7 | 7.2 | |
| Albumin | ILI (COVID-19 negative) | 3.847 | 3.9 | 0.349 | 0 | 3.1 | 1.5 | 4.6 | 3.9 | 3.9 | 3.9 |
| COVID-19 positive | 3.846 | 3.9 | 0.574 | 0.9 | 6.6 | 0.4 | 7 | 3.4 | 3.9 | 4.3 | |
| HbA1c | ILI (COVID-19 negative) | 6.1 | 5.8 | 1.311 | 0 | 9.1 | 4 | 13.1 | 5.8 | 5.8 | 5.8 |
| COVID-19 positive | 6.806 | 6.2 | 1.872 | 1.8 | 14.2 | 4 | 18.2 | 5.6 | 6.2 | 7.4 |
| Algorithm | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) | AUC (%) |
|---|---|---|---|---|---|
| Random forest | 94 | 94 | 89 | 91 | 99 |
| Logistic regression | 68 | 65 | 70 | 68 | 74 |
| Decision tree | 81 | 75 | 83 | 77 | 88 |
| KNN | 81 | 75 | 83 | 77 | 83 |
| STACKA | 90 | 85 | 90 | 87 | 96 |
| Adaboost | 94 | 91 | 94 | 92 | 95 |
| Catboost | 90 | 86 | 86 | 86 | 96 |
| Lightgbm | 96 | 94 | 95 | 94 | 98 |
| Xgboost | 96 | 95 | 93 | 94 | 99 |
| STACKB | 96 | 95 | 95 | 95 | 99 |
| STACKC | 96 | 94 | 95 | 94 | 98 |
| Algorithm | Hyperparameters Chosen |
|---|---|
| Random forest | {‘bootstrap’: True, ‘max_depth’: 110, ‘max_features’: 2, ‘min_samples_leaf’: 3, ‘min_samples_split’: 8, ‘n_estimators’: 300} |
| Logistic regression | {‘C’: 100, ‘penalty’: ‘l2’} |
| Decision tree | {‘criterion’: ‘gini’, ‘max_depth’: 40, ‘max_features’: ‘sqrt’, ‘min_samples_leaf’: 1, ‘min_samples_split’: 10, ‘splitter’: ‘best’} |
| KNN | {‘n_neighbors’: 1} |
| STACKA | {use_probas=True, average_probas=False, meta_classifier=Logistic Regresion} |
| Adaboost | {‘learning_rate’: 1.0, ‘n_estimators’: 300} |
| Catboost | {‘border_count’: 32, ‘depth’: 3, ‘iterations’: 250, ‘l2_leaf_reg’: 3, ‘learning_rate’: 0.03} |
| Lightgbm | {‘lambda_l1’: 0, ‘lambda_l2’: 1, ‘min_data_in_leaf’: 30, ‘num_leaves’: 31, ‘reg_alpha’: 0.1} |
| Xgboost | {‘colsample_bytree’: 0.3, ‘gamma’: 0.0, ‘learning_rate’: 0.1, ‘max_depth’: 8, ‘min_child_weight’: 1} |
| STACKB | {use_probas=True, average_probas=False, meta_classifier=Logistic Regresion} |
| STACKC | {use_probas=True, average_probas=False, meta_classifier=Logistic Regresion} |
| Model: “Sequential” | ||
|---|---|---|
| Layer (type) | Output shape | Parameters |
| dense (Dense) | (none, 21) | 462 |
| dense_1 (Dense) | (none, 12) | 264 |
| dense_2 (Dense) | (none, 9) | 117 |
| dense_3 (Dense) | (none, 7) | 70 |
| dense_4 (Dense) | (none, 4) | 32 |
| dense_5 (Dense) | (none, 1) | 5 |
| Total parameters: 950 | ||
| Trainable parameters:950 | ||
| Non-trainable parameters:950 |
| Model: “Sequential” | ||
|---|---|---|
| Layer (type) | Output Shape | Parameters |
| conv1d (Conv1D) | (none, 21, 32) | 128 |
| conv1d_1 (Conv1D) | (none, 21, 64) | 6208 |
| conv1d_2 (Conv1D) | (none, 21, 128) | 24,704 |
| max_pooling1d (MaxPooling1D) | (none, 11, 128) | 0 |
| dropout (Dropout) | (none, 11, 128) | 0 |
| flatten (Flatten) | (none, 1408) | 0 |
| dense (Dense) | (none, 256) | 360,704 |
| dense_1 (Dense) | (none, 512) | 131,584 |
| dense_2 (Dense) | (none, 1) | 513 |
| Total params: 523,841 | ||
| Trainable params: 523,841 | ||
| Non-trainable params: 0 |
| Deep Learning Model | Accuracy (in %) | Precision (in %) | Recall (in %) | F1-Score (in %) | AUC (in %) | Hyperparameters |
|---|---|---|---|---|---|---|
| DNN | 87 | 80 | 86 | 83 | 90 | Number of layers: six, neurons: (21,12,9,7,4,1), activation function: relu for first five layers and sigmoid for the last layer, optimizer: adam, loss function: binary cross entropy, batch size: 10, epochs: 1000, learning rate: 0.0001 |
| 1D-CNN | 90 | 86 | 89 | 88 | 93 | Number of layers: nine, activation function: leaky relu for first eight layers and sigmoid for the last layer, optimizer: adam, loss function: binary cross entropy, batch size: 10, epochs: 200, learning rate: 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chadaga, K.; Prabhu, S.; Bhat, V.; Sampathila, N.; Umakanth, S.; Chadaga, R. A Decision Support System for Diagnosis of COVID-19 from Non-COVID-19 Influenza-like Illness Using Explainable Artificial Intelligence. Bioengineering 2023, 10, 439. https://doi.org/10.3390/bioengineering10040439
Chadaga K, Prabhu S, Bhat V, Sampathila N, Umakanth S, Chadaga R. A Decision Support System for Diagnosis of COVID-19 from Non-COVID-19 Influenza-like Illness Using Explainable Artificial Intelligence. Bioengineering. 2023; 10(4):439. https://doi.org/10.3390/bioengineering10040439
Chicago/Turabian StyleChadaga, Krishnaraj, Srikanth Prabhu, Vivekananda Bhat, Niranjana Sampathila, Shashikiran Umakanth, and Rajagopala Chadaga. 2023. "A Decision Support System for Diagnosis of COVID-19 from Non-COVID-19 Influenza-like Illness Using Explainable Artificial Intelligence" Bioengineering 10, no. 4: 439. https://doi.org/10.3390/bioengineering10040439
APA StyleChadaga, K., Prabhu, S., Bhat, V., Sampathila, N., Umakanth, S., & Chadaga, R. (2023). A Decision Support System for Diagnosis of COVID-19 from Non-COVID-19 Influenza-like Illness Using Explainable Artificial Intelligence. Bioengineering, 10(4), 439. https://doi.org/10.3390/bioengineering10040439