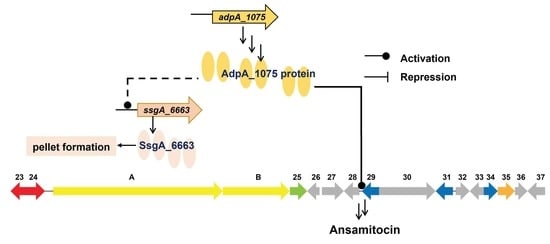
Global Regulator AdpA_1075 Regulates Morphological Differentiation and Ansamitocin Production in Actinosynnema pretiosum subsp. auranticum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids and Culture Conditions
2.2. Construction of Recombinant Strains
2.3. RNA Isolation, cDNA Synthesis and Quantitative Real-Time PCR (qRT-PCR)
2.4. Determination of AP-3 Production
2.5. Mycelial Morphology Observation
2.6. Scanning Electron Microscope (SEM)
2.7. Heterologous Overexpression of AdpA-1075
2.8. Electrophoretic Mobility Shift Assays (EMSA)
3. Results
3.1. Identification of ssgA in A. pretiosum subsp. auranticum
3.2. Deletion of ssgA_6663 Affected the Morphological Differentiation of A. pretiosum
3.3. Overexpression of adpA_1075 Increased the Production of AP-3
3.4. AdpA_1075 Is Involved in the Regulation of ssgA_6663 Transcription in A. pretiosum
3.5. AdpA_1075 Binds to Promoters of ssgA_6663 and asm28
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Prelog, V.; Oppolzer, W. Ansamycins, a novel class of microbial metabolites. Helv. Chim. Acta 1973, 56, 2279–2287. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.; Müller, P.; Schreiner, J.; Prince, S.S.; Lardinois, D.; Heinzelmann-Schwarz, V.A.; Thommen, D.S.; Zippelius, A. The microtubule-depolymerizing agent ansamitocin P3 programs dendritic cells toward enhanced anti-tumor immunity. Cancer Immunol. Immunother 2014, 63, 925–938. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, A.S.; Fernandez-Rodriguez, L.; Zhao, Y.; Monaco, G.; Trefny, M.P.; Yoshida, N.; Martin, K.; Sharma, A.; Olieric, N.; Shah, P.; et al. GEF-H1 signaling upon microtubule destabilization is required for dendritic cell activation and specific anti-tumor responses. Cell Rep. 2019, 28, 3367–3380.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barok, M.; Joensuu, H.; Isola, J. Trastuzumab emtansine: Mechanisms of action and drug resistance. Breast Cancer Res. 2014, 16, 209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, Y.; Gao, Y.; Zhou, J.; Wei, L.; Chen, J.; Hua, Q. Process optimization with alternative carbon sources and modulation of secondary metabolism for enhanced ansamitocin P-3 production in Actinosynnema pretiosum. J. Biotechnol. 2014, 192, 1–10. [Google Scholar] [CrossRef]
- Li, T.; Fan, Y.; Nambou, K.; Hu, F.; Imanaka, T.; Wei, L.; Hua, Q. Improvement of ansamitocin P-3 production by Actinosynnema mirum with fructose as the sole carbon source. Appl. Biochem. Biotechnol. 2015, 175, 2845–2856. [Google Scholar] [CrossRef]
- Fan, Y.; Hu, F.; Wei, L.; Bai, L.; Hua, Q. Effects of modulation of pentose-phosphate pathway on biosynthesis of ansamitocins in Actinosynnema pretiosum. J. Biotechnol. 2016, 230, 3–10. [Google Scholar] [CrossRef]
- Zhao, M.; Fan, Y.; Wei, L.; Hu, F.; Hua, Q. Effects of the methylmalonyl-CoA metabolic pathway on ansamitocin production in Actinosynnema pretiosum. Appl. Biochem. Biotechnol. 2017, 181, 1167–1178. [Google Scholar] [CrossRef]
- Ning, X.; Wang, X.; Wu, Y.; Kang, Q.; Bai, L. Identification and engineering of post-PKS modification bottlenecks for ansamitocin P-3 titer improvement in Actinosynnema pretiosum subsp. pretiosum ATCC 31280. Biotechnol. J. 2017, 12, 1700484. [Google Scholar] [CrossRef]
- Du, Z.Q.; Zhang, Y.; Qian, Z.G.; Xiao, H.; Zhong, J.J. Combination of traditional mutation and metabolic engineering to enhance ansamitocin P-3 production in Actinosynnema pretiosum. Biotechnol. Bioeng. 2017, 114, 2794–2806. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.Q.; Zhong, J.J. Rational approach to improve ansamitocin P-3 production by integrating pathway engineering and substrate feeding in Actinosynnema pretiosum. Biotechnol. Bioeng. 2018, 115, 2456–2466. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Guo, S.; Hua, Q.; Hu, F. Improved AP-3 production through combined ARTP mutagenesis, fermentation optimization, and subsequent genome shuffling. Biotechnol. Lett. 2021, 43, 1143–1154. [Google Scholar] [CrossRef]
- Kumar, P.; Dubey, K.K. Mycelium transformation of Streptomyces toxytricini into pellet: Role of culture conditions and kinetics. Bioresour. Technol. 2017, 228, 339–347. [Google Scholar] [CrossRef]
- Celler, K.; Picioreanu, C.; van Loosdrecht, M.C.M.; van Wezel, G.P. Structured morphological modeling as a framework for rational strain design of Streptomyces species. Antonie Van Leeuwenhoek 2012, 102, 409–423. [Google Scholar] [CrossRef] [Green Version]
- Paul, G.C.; Thomas, C.R. Characterisation of mycelial morphology using image analysis. Adv. Biochem. Eng. Biotechnol. 1998, 60, 1–59. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhao, G.; Ding, X. Morphology engineering of Streptomyces coelicolor M145 by sub-inhibitory concentrations of antibiotics. Sci. Rep. 2017, 7, 13226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, A.; Pierson, D.L.; Mishra, S.K.; Demain, A.L. Growth of Streptomyces hygroscopicus in rotating-wall bioreactor under simulated microgravity inhibits rapamycin production. Appl. Microbiol. Biotechnol. 2000, 54, 33–36. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.; Tamura, S.; Koike, Y.; Toriyama, M.; Okabe, M. Mycelial pellet intrastructure visualization and viability prediction in a culture of Streptomyces fradiae using confocal scanning laser microscopy. J. Ferment. Bioeng. 1997, 84, 483–486. [Google Scholar] [CrossRef]
- Jonsbu, E.; McIntyre, M.; Nielsen, J. The influence of carbon sources and morphology on nystatin production by Streptomyces noursei. J. Biotechnol. 2002, 95, 133–144. [Google Scholar] [CrossRef]
- Vecht-Lifshitz, S.E.; Sasson, Y.; Braun, S. Nikkomycin production in pellets of Streptomyces tendae. J. Appl. Bacteriol. 1992, 72, 195–200. [Google Scholar] [CrossRef]
- Wardell, J.N.; Stocks, S.M.; Thomas, C.R.; Bushell, M.E. Decreasing the hyphal branching rate of Saccharopolyspora erythraea NRRL 2338 leads to increased resistance to breakage and increased antibiotic production. Biotechnol. Bioeng. 2002, 78, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Sarrà, M.; Casas, C.; Poch, M.; Gòdia, F. A simple structured model for continuous production of a hybrid antibiotic by Streptomyces lividans pellets in a fluidized-bed bioreactor. Appl. Biochem. Biotechnol. 1999, 80, 39–50. [Google Scholar] [CrossRef]
- van Dissel, D.; Claessen, D.; van Wezel, G.P. Chapter One-Morphogenesis of Streptomyces in submerged cultures. Adv. Appl. Microbiol. 2014, 89, 1–45. [Google Scholar] [PubMed]
- Noens, E.E.; Mersinias, V.; Willemse, J.; Traag, B.A.; Laing, E.; Chater, K.F.; Smith, C.P.; Koerten, H.K.; Van Wezel, G.P. Loss of the controlled localization of growth stage-specific cell-wall synthesis pleiotropically affects developmental gene expression in an ssgA mutant of Streptomyces coelicolor. Mol. Microbiol. 2007, 64, 1244–1259. [Google Scholar] [CrossRef]
- Nothaft, H.; Dresel, D.; Willimek, A.; Mahr, K.; Niederweis, M.; Titgemeyer, F. The phosphotransferase system of Streptomyces coelicolor is biased for N-acetylglucosamine metabolism. J. Bacteriol. 2003, 185, 7019–7023. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.; Kendrick, K.E. Characterization of ssfR and ssgA, two genes involved in sporulation of Streptomyces griseus. J. Bacteriol. 2000, 182, 5521–5529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Wezel, G.P.; van der Meulen, J.; Kawamoto, S.; Luiten, R.G.M.; Koerten, H.K.; Kraal, B. SsgA Is Essential for Sporulation of Streptomyces Coelicolor A3(2) and Affects Hyphal Development by Stimulating Septum Formation. J. Bacteriol. 2000, 182, 5653–5662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamazaki, H.; Ohnishi, Y.; Horinouchi, S. Transcriptional Switch on of SsgA by A-Factor, Which Is Essential for Spore Septum Formation in Streptomyces Griseus. J. Bacteriol. 2003, 185, 1273–1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bi, E.; Lutkenhaus, J. FtsZ Ring Structure Associated with Division in Escherichia Coli. Nature 1991, 354, 161–164. [Google Scholar] [CrossRef]
- Xu, W.; Huang, J.; Lin, R.; Shi, J.; Cohen, S.N. Regulation of Morphological Differentiation in S. Coelicolor by RNase III (AbsB) Cleavage of MRNA Encoding the AdpA Transcription Factor: AbsB Regulates AdpA in S. Coelicolor. Mol. Microbiol. 2010, 75, 781–791. [Google Scholar] [CrossRef]
- Xiao, X.; Willemse, J.; Voskamp, P.; Li, X.; Prota, A.E.; Lamers, M.; Pannu, N.; Abrahams, J.P.; van Wezel, G.P. Ectopic Positioning of the Cell Division Plane Is Associated with Single Amino Acid Substitutions in the FtsZ-Recruiting SsgB in Streptomyces. Open. Biol. 2021, 11, 200409. [Google Scholar] [CrossRef] [PubMed]
- Van Wezel, G.P.; Krabben, P.; Traag, B.A.; Keijser, B.J.; Kerste, R.; Vijgenboom, E.; Heijnen, J.J.; Kraal, B. Unlocking Streptomyces spp. for use as sustainable industrial production platforms by morphological engineering. Appl. Environ. Microbiol. 2006, 72, 5283–5288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, H.T.; Pham, V.T.T.; Nguyen, C.T.; Pokhrel, A.R.; Kim, T.-S.; Kim, D.; Na, K.; Yamaguchi, T.; Sohng, J.K. Exploration of cryptic organic photosensitive compound as zincphyrin IV in Streptomyces venezuelae ATCC 15439. Appl. Microbiol. Biotechnol. 2020, 104, 713–724. [Google Scholar] [CrossRef]
- Traag, B.A.; Kelemen, G.H.; Van Wezel, G.P. Transcription of the sporulation gene ssgA is activated by the IclR-type regulator SsgR in a whi-independent manner in Streptomyces coelicolor A3(2). Mol. Microbiol. 2004, 53, 985–1000. [Google Scholar] [CrossRef] [Green Version]
- Horinouchi, S.; Beppu, T. Hormonal control by A-factor of morphological development and secondary metabolism in Streptomyces. Proc. Jpn. Acad. Ser. B 2007, 83, 277–295. [Google Scholar] [CrossRef] [Green Version]
- Ohnishi, Y.; Kameyama, S.; Onaka, H.; Horinouchi, S. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: Identification of a target gene of the A-factor receptor. Mol. Microbiol. 1999, 34, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, Y.; Yamazaki, H.; Kato, J.; Tomono, A.; Horinouchi, S. AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci. Biotechnol. Biochem. 2005, 69, 431–439. [Google Scholar] [CrossRef] [Green Version]
- Akanuma, G.; Hara, H.; Ohnishi, Y.; Horinouchi, S. Dynamic changes in the extracellular proteome caused by absence of a pleiotropic regulator AdpA in Streptomyces griseus. Mol. Microbiol. 2009, 73, 898–912. [Google Scholar] [CrossRef] [PubMed]
- Bush, M.J.; Tschowri, N.; Schlimpert, S.; Flärdh, K.; Buttner, M.J. c-di-GMP signalling and the regulation of developmental transitions in streptomycetes. Nat. Rev. Microbiol. 2015, 13, 749–760. [Google Scholar] [CrossRef]
- Bu, X.-L.; Weng, J.-Y.; He, B.-B.; Xu, M.-J.; Xu, J. A novel AdpA homologue negatively regulates morphological differentiation in Streptomyces xiamenensis 318. Appl. Environ. Microbiol. 2019, 85, e03107-18. [Google Scholar] [CrossRef]
- Romero-Rodríguez, A.; Robledo-Casados, I.; Sánchez, S. An overview on transcriptional regulators in Streptomyces. Biochim. Biophys. Acta 2015, 1849, 1017–1039. [Google Scholar] [CrossRef] [PubMed]
- Higo, A.; Hara, H.; Horinouchi, S.; Ohnishi, Y. Genome-wide distribution of AdpA, a global regulator for secondary metabolism and morphological differentiation in Streptomyces, revealed the extent and complexity of the AdpA regulatory network. DNA Res. 2012, 19, 259–273. [Google Scholar] [CrossRef]
- Rabyk, M.; Yushchuk, O.; Rokytskyy, I.; Anisimova, M.; Ostash, B. Genomic insights into evolution of AdpA family master regulators of morphological differentiation and secondary metabolism in Streptomyces. J. Mol. Evol. 2018, 86, 204–215. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Kang, Q.; Zhang, L.-L.; Bai, L. Subtilisin-involved morphology engineering for improved antibiotic production in actinomycetes. Biomolecules 2020, 10, 851. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, R.; Kang, Q.; Bai, L. The antitumor agent ansamitocin P-3 binds to cell division protein FtsZ in Actinosynnema pretiosum. Biomolecules 2020, 10, 699. [Google Scholar] [CrossRef]
- Zhang, P.; Zhang, K.; Liu, Y.; Fu, J.; Zong, G.; Ma, X.; Cao, G. Deletion of the response regulator PhoP accelerates the formation of aerial mycelium and spores in Actinosynnema pretiosum. Front. Microbiol. 2022, 13, 845620. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Sun, X.; Li, R.; Zhang, T.; Hu, F.; Liu, F.; Hua, Q. Two strategies to improve the supply of pks extender units for ansamitocin P-3 biosynthesis by CRISPR–Cas9. Bioresour. Bioprocess. 2022, 9, 90. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Xu, H.; Chater, K.F.; Deng, Z.; Tao, M. A cellulose synthase-like protein involved in hyphal tip growth and morphological differentiation in Streptomyces. J. Bacteriol. 2008, 190, 4971–4978. [Google Scholar] [CrossRef] [Green Version]
- Kawamoto, S.; Watanabe, H.; Hesketh, A.; Ensign, J.C.; Ochi, K. Expression analysis of the ssgA gene product, associated with sporulation and cell division in Streptomyces griseus. Microbiology 1997, 143, 1077–1086. [Google Scholar] [CrossRef]
- McCormick, J.R.; Su, E.P.; Driks, A.; Losick, R. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol. Microbiol. 1994, 14, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Santos-Beneit, F.; Roberts, D.M.; Cantlay, S.; McCormick, J.R.; Errington, J. A mechanism for FtsZ-independent proliferation in Streptomyces. Nat. Commun. 2017, 8, 1378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yushchuk, O.; Ostash, I.; Vlasiuk, I.; Gren, T.; Luzhetskyy, A.; Kalinowski, J.; Fedorenko, V.; Ostash, B. Heterologous AdpA transcription factors enhance landomycin production in Streptomyces cyanogenus S136 under a broad range of growth conditions. Appl. Microbiol. Biotechnol. 2018, 102, 8419–8428. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Wang, Y.; Hou, B.; Wang, R.; Ye, J.; Zhu, X.; Wu, H.; Zhang, H. AdpAlin, a pleiotropic transcriptional regulator, is involved in the cascade regulation of lincomycin biosynthesis in Streptomyces lincolnensis. Front. Microbiol. 2019, 10, 2428. [Google Scholar] [CrossRef] [PubMed]
- Higo, A.; Horinouchi, S.; Ohnishi, Y. Strict regulation of morphological differentiation and secondary metabolism by a positive feedback loop between two global regulators AdpA and BldA in Streptomyces griseus. Mol. Microbiol. 2011, 81, 1607–1622. [Google Scholar] [CrossRef] [PubMed]
- Guyet, A.; Benaroudj, N.; Proux, C.; Gominet, M.; Coppée, J.-Y.; Mazodier, P. Identified members of the Streptomyces lividans AdpA regulon involved in differentiation and secondary metabolism. BMC Microbiol. 2014, 14, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamazaki, H.; Tomono, A.; Ohnishi, Y.; Horinouchi, S. DNA-binding specificity of AdpA, a transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus: DNA-binding specificity of AdpA. Mol. Microbiol. 2004, 53, 555–572. [Google Scholar] [CrossRef]
- Watanabe, K.; Okuda, T.; Yokose, K.; Furumai, T.; Maruyama, H. Actinosynnema mirum, a new producer of nocardicin antibiotics. J. Antibiot. 1983, 36, 321–324. [Google Scholar] [CrossRef] [PubMed]
- Keijser, B.J.F.; Noens, E.E.E.; Kraal, B.; Koerten, H.K.; van Wezel, G.P. The Streptomyces coelicolor ssgB gene is required for early stages of sporulation. FEMS Microbiol. Lett. 2003, 225, 59–67. [Google Scholar] [CrossRef] [Green Version]
- Xu, Q.; Traag, B.A.; Willemse, J.; McMullan, D.; Miller, M.D.; Elsliger, M.-A.; Abdubek, P.; Astakhova, T.; Axelrod, H.L.; Bakolitsa, C.; et al. Structural and functional characterizations of SsgB, a conserved activator of developmental cell division in morphologically complex actinomycetes. J. Biol. Chem. 2009, 284, 25268–25279. [Google Scholar] [CrossRef] [PubMed]
- Makitrynskyy, R.; Ostash, B.; Tsypik, O.; Rebets, Y.; Doud, E.; Meredith, T.; Luzhetskyy, A.; Bechthold, A.; Walker, S.; Fedorenko, V. Pleiotropic regulatory genes bldA, adpA and absB are implicated in production of phosphoglycolipid antibiotic moenomycin. Open Biol. 2013, 3, 130121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bandi, S.; Kim, Y.; Chang, Y.K.; Shang, G.; Yu, T.W.; Floss, H.G. Construction of asm2 deletion mutant of Actinosynnema pretiosum and medium optimization for ansamitocin P-3 production using statistical approach. J. Microbiol. Biotechnol. 2006, 16, 1338–1346. [Google Scholar]
- Ng, D.; Chin, H.K.; Wong, V.V.T. Constitutive overexpression of asm2 and asm39 increases AP-3 production in the actinomycete Actinosynnema pretiosum. J. Ind. Microbiol. Biotechnol. 2009, 36, 1345–1351. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Kang, Q.; Wang, L.; Bai, L.; Deng, Z. Asm8, a specific LAL-type activator of 3-amino-5-hydroxybenzoate biosynthesis in ansamitocin production. Sci. China Life Sci. 2013, 56, 601–608. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Lu, C.; Chang, X.; Shen, Y. Constitutive overexpression of asm18 increases the production and diversity of maytansinoids in Actinosynnema pretiosum. Appl. Microbiol. Biotechnol. 2016, 100, 2641–2649. [Google Scholar] [CrossRef]
- Hackl, S.; Bechthold, A. The gene bldA, a regulator of morphological differentiation and antibiotic production in Streptomyces. Arch. Pharm. 2015, 348, 455–462. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, S.; Leng, T.; Sun, X.; Zheng, J.; Li, R.; Chen, J.; Hu, F.; Liu, F.; Hua, Q. Global Regulator AdpA_1075 Regulates Morphological Differentiation and Ansamitocin Production in Actinosynnema pretiosum subsp. auranticum. Bioengineering 2022, 9, 719. https://doi.org/10.3390/bioengineering9110719
Guo S, Leng T, Sun X, Zheng J, Li R, Chen J, Hu F, Liu F, Hua Q. Global Regulator AdpA_1075 Regulates Morphological Differentiation and Ansamitocin Production in Actinosynnema pretiosum subsp. auranticum. Bioengineering. 2022; 9(11):719. https://doi.org/10.3390/bioengineering9110719
Chicago/Turabian StyleGuo, Siyu, Tingting Leng, Xueyuan Sun, Jiawei Zheng, Ruihua Li, Jun Chen, Fengxian Hu, Feng Liu, and Qiang Hua. 2022. "Global Regulator AdpA_1075 Regulates Morphological Differentiation and Ansamitocin Production in Actinosynnema pretiosum subsp. auranticum" Bioengineering 9, no. 11: 719. https://doi.org/10.3390/bioengineering9110719
APA StyleGuo, S., Leng, T., Sun, X., Zheng, J., Li, R., Chen, J., Hu, F., Liu, F., & Hua, Q. (2022). Global Regulator AdpA_1075 Regulates Morphological Differentiation and Ansamitocin Production in Actinosynnema pretiosum subsp. auranticum. Bioengineering, 9(11), 719. https://doi.org/10.3390/bioengineering9110719