A Comparative Study of Morphology, Photosynthetic Physiology, and Proteome between Diploid and Tetraploid Watermelon (Citrullus lanatus L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
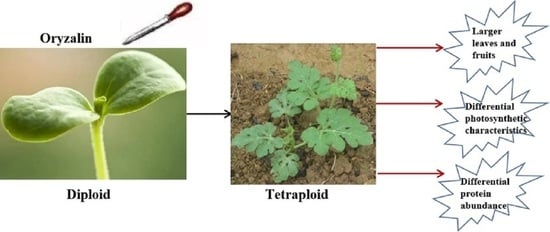
2.2. Tetraploid Watermelon Induction by Oryzalin
2.3. Polyploid Watermelon Chromosome Counting
2.4. Flow Cytometry
2.5. Guard Cell Number and Chloroplast Number in Guard Cells
2.6. Determination of Leaf Morphology
2.7. Photosynthesis and Chlorophyll Content
2.8. Watermelon Leaf Protein Extraction
2.9. Two-Dimensional Gel Electrophoresis of Proteins
2.10. Protein Analysis by MALDI-TOF-MS/MS
2.11. Generation of Protein–Protein Interaction Networks
2.12. RNA Isolation and qRT-PCR
2.13. Statistical Analysis
3. Results
3.1. Confirmation of Watermelon Polyploidy
3.2. Morphological Comparison of Diploid and Tetraploid Watermelon
3.3. Physiological Analysis of Diploid and Tetraploid Watermelon
3.4. Protein Profile of Diploid and Tetraploid Watermelon
3.5. Functional Analysis of Screened Proteins
3.6. Protein Interaction Networks
3.7. Validation of Differentially Expressed Proteins by qRT-PCR
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Garcia-Lozano, M.; Dutta, S.K.; Natarajan, P.; Tomason, Y.R.; Lopez, C.; Katam, R.; Levi, A.; Nimmakayala, P.; Reddy, U.K. Transcriptome changes in reciprocal grafts involving watermelon and bottle gourd reveal molecular mechanisms involved in increase of the fruit size, rind toughness and soluble solids. Plant Mol. Biol. 2020, 102, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Niazi, I.A.K.; Rauf, S.; Teixeira da Silva, J.A.; Iqbal, Z.; Munir, H. Induced polyploidy in inter-subspecific maize hybrids to reduce heterosis breakdown and restore reproductive fertility. Grass Forage Sci. 2015, 70, 682–694. [Google Scholar] [CrossRef]
- Dabkevičienė, G.; Statkevičiūtė, G.; Mikaliūnienė, J.; Norkevičienė, E.; Kemešytė, V. Production of Trifolium pratense L. and T. hybridum L. tetraploid populations and assessment of their agrobiological characteristics. Zemdirb. Agric. 2016, 103, 377–384. [Google Scholar] [CrossRef] [Green Version]
- Luo, Z.N.; Iaffaldano, B.J.; Cornish, K. Colchicine-induced polyploidy has the potential to improve rubber yield in Taraxacum kok-saghyz. Ind. Crops Prod. 2018, 112, 75–81. [Google Scholar] [CrossRef]
- Rao, S.P.; Tian, Y.R.; Xia, X.L.; Li, Y.; Chen, J.H. Chromosome doubling mediates superior drought tolerance in Lycium ruthenicum via abscisic acid signaling. Hortic. Res. 2020, 7, 40. [Google Scholar] [CrossRef] [Green Version]
- Khalid, M.F.; Hussain, S.; Anjum, M.A.; Ahmad, S.; Ali, M.A.; Ejaz, S.; Morillon, R. Better salinity tolerance in tetraploid vs diploid volkamer lemon seedlings is associated with robust antioxidant and osmotic adjustment mechanisms. J. Plant Physiol. 2020, 244, 153071. [Google Scholar] [CrossRef]
- Oustric, J.; Morillon, R.; Luro, F.; Herbette, S.; Martin, P.; Giannettini, J.; Berti, L.; Santini, J. Nutrient Deficiency Tolerance in Citrus Is Dependent on Genotype or Ploidy Level. Front. Plant Sci. 2019, 11, 127. [Google Scholar] [CrossRef]
- Jaskani, M.J.; Kwon, S.W.; Kim, D.H. Comparative study on vegetative, reproductive and qualitative traits of seven diploid and tetraploid watermelon lines. Euphytica 2005, 145, 259–268. [Google Scholar] [CrossRef]
- Zhu, H.J.; Zhao, S.J.; Lu, X.Q.; He, N.; Gao, L.; Dou, J.L.; Bie, Z.L.; Liu, W.G. Genome duplication improves the resistance of watermelon root to salt stress. Plant Physiol. Biochem. 2018, 133, 11–21. [Google Scholar] [CrossRef]
- Zhang, N.; Bao, Y.N.; Xie, Z.L.; Huang, X.; Sun, Y.H.; Feng, G.; Zeng, H.X.; Ren, J.; Li, Y.H.; Xiong, J.S.; et al. Efficient Characterization of Tetraploid Watermelon. Plants 2019, 8, 419. [Google Scholar] [CrossRef]
- Liu, G.F.; Li, Z.N.; Bao, M.Z. Colchicine-induced chromosome doubling in Platanus acerifolia and its effect on plant morphology. Euphytica 2007, 157, 145–154. [Google Scholar] [CrossRef]
- Sari, N.; Abak, K.; Pitrat, M. Comparison of ploidy level screening methods in watermelon: Citrullus lanatus (Thunb.) Matsum. and Nakai. Sci. Hortic. 1999, 82, 265–277. [Google Scholar] [CrossRef]
- Chen, S.B.; Gollop, N.; Heuer, B. Proteomic analysis of salt-stressed tomato (Solanum lycposersicum) seedlings: Effect of genotype and exogenous application of glycinebetaine. J. Exp. Bot. 2009, 60, 2005–2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, H.F.; Xu, Y.L.; Du, C.X.; Wu, X. Phloem sap proteome studied by iTRAQ provides integrated insight into salinity response mechanisms in cucumber plants. J. Proteom. 2015, 125, 54–67. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Zhang, M.; Xu, J.H.; Liu, G.; Yao, X.F.; Ren, R.S.; Yang, X.P. Proteomic analysis of responsive root proteins of Fusarium oxysporum-infected watermelon seedlings. Plant Soil 2018, 422, 169–181. [Google Scholar] [CrossRef]
- Arndt, C.; Koristka, S.; Feldmann, A.; Bartsch, H.; Bachmann, M. Coomassie-Brilliant blue staining of polyacrylamide gels. Methods Mol. Biol. 2012, 869, 465–469. [Google Scholar]
- Domínguez-Delgado, J.J.; López-Jurado, J.; Mateos-Naranjo, E.; Balao, F. Phenotypic diploidization in plant functional traits uncovered by synthetic neopolyploids in Dianthus broteri. J. Exp. Bot. 2021, 72, 5522–5533. [Google Scholar] [CrossRef]
- Bogunić, F.; Siljak-Yakovlev, S.; Mahmutović-Dizdarević, I.; Hajrudinović-Bogunić, A.; Bourge, M.; Brown, S.C.; Muratović, E. Genome Size, Cytotype Diversity and Reproductive Mode Variation of Cotoneaster integerrimus (Rosaceae) from the Balkans. Plants 2021, 10, 2798. [Google Scholar] [CrossRef]
- Pei, Y.; Yao, N.; He, L.; Deng, D.X.; Li, W.; Zhang, W.P. Comparative study of the morphological, physiological and molecular characteristics between diploid and tetraploid radish (Raphunas sativus L.). Sci. Hortic. 2019, 257, 108739. [Google Scholar] [CrossRef]
- Marzougui, N.; Boubaya, A.; Thabti, I.; Elfalleh, W.; Guasmi, F.; Ferchichi, A. Polyploidy induction of Tunisian Trigonella foenumgreaum L. populations. Afr. J. Biotechnol. 2011, 10, 8570–8577. [Google Scholar]
- Pinheiro, A.A.; Pozzobon, M.T.; do Valle, C.B.; Penteado, M.I.O.; Carneiro, V.T.C. Duplication of the chromosome number of diploid Brachiaria brizantha plants using colchicine. Plant Cell Rep. 2000, 19, 274–278. [Google Scholar] [CrossRef] [PubMed]
- Saisingtong, S.; Schmid, J.E.; Stamp, P.; Büter, B. Colchicine-mediated chromosome doubling during another culture of maize (Zea mays L.). Theor. Appl. Genet. 1996, 92, 1017–1023. [Google Scholar] [CrossRef] [PubMed]
- Noh, J.; Sheikh, S.; Chon, H.G.; Seong, M.H.; Lim, J.H.; Lee, S.G.; Jung, G.T.; Kim, J.M.; Ju, H.J.; Huh, Y.C. Screening different methods of tetraploid induction in watermelon [Citrullus lanatus (thunb.) Manst. and Nakai]. Hortic. Environ. Biotechnol. 2012, 53, 521–529. [Google Scholar] [CrossRef]
- Hansen, N.J.P.; Andersen, S.B. In vitro chromosome doubling potential of colchicine, oryzalin, trifluralin, and APM in Brassica napus microspore culture. Euphytica 1996, 88, 159–164. [Google Scholar] [CrossRef]
- Hooghvorst, I.; Nogués, S. Opportunities and Challenges in Doubled Haploids and Haploid Inducer-Mediated Genome-Editing Systems in Cucurbits. Agronomy 2020, 10, 1441. [Google Scholar] [CrossRef]
- Abdoli, M.; Moieni, A.; Badi, H.N. Morphological, physiological, cytological and phytochemical studies in diploid and colchicine-induced tetraploid plants of Echinacea purpurea (L.). Acta Physiol. Plant. 2013, 35, 2075–2083. [Google Scholar] [CrossRef]
- El-Morsy, S.I.; Dorra, M.D.M.; Abd El-Hady, E.A.A.; Hiaba, A.A.A.; Mohamed, A.Y. Comparative Studies on Diploid and Tetraploid Levels of Nicotiana alata. Acad. J. Plant Sci. 2009, 2, 182–188. [Google Scholar]
- Tang, Z.Q.; Chen, D.L.; Song, Z.J.; He, Y.C.; Cai, D.T. In vitro induction and identification of tetraploid plants of Paulownia tomentosa. Plant Cell Tissue Organ Cult. 2010, 102, 213–220. [Google Scholar] [CrossRef]
- Sabzehzari, M.; Hoveidamanesh, S.; Modarresi, M.; Mohammadi, V. Morphological, anatomical, physiological, and cytological studies in diploid and tetraploid plants of Plantago psyllium. Plant Cell Tissue Organ Cult. 2019, 139, 131–137. [Google Scholar] [CrossRef]
- Saminathan, T.; Nimmakayala, P.; Manohar, S.; Malkaram, S.; Almeida, A.; Cantrell, R.; Tomason, Y.; Abburi, L.; Rahman, M.A.; Vajja, V.G.; et al. Differential gene expression and alternative splicing between diploid and tetraploid watermelon. J. Exp. Bot. 2015, 66, 1369–1385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Lozano, M.; Levi, A.; Katam, R.; Lopez-Ortiz, C.; Nimmakayala, P.; Reddy, U.K. Altered chromatin conformation and transcriptional regulation in watermelon following genome doubling. Plant J. 2021, 106, 588–600. [Google Scholar] [CrossRef] [PubMed]
- Dou, J.L.; Yuan, P.L.; Zhao, S.J.; He, N.; Zhu, H.J.; Gao, L.; Ji, W.L.; Lu, X.Q.; Liu, W.G. Effect of ploidy level on expression of lycopene biosynthesis genes and accumulation of phytohormones during watermelon (Citrullus lanatus) fruit development and ripening. J. Integr. Agric. 2017, 16, 1956–1967. [Google Scholar] [CrossRef]
- Wang, Y.; Jia, B.Y.; Ren, H.J.; Feng, Z. Ploidy level enhances the photosynthetic capacity of a tetraploid variety of Acer buergerianum Miq. Peer J. 2021, 9, e12620. [Google Scholar] [CrossRef]
- Liu, S.Y.; Chen, S.M.; Chen, Y.; Guan, Z.Y.; Yin, D.M.; Chen, F.D. In vitro induced tetraploid of Dendranthema nankingense (Nakai) Tzvel. shows an improved level of abiotic stress tolerance. Sci. Hortic. 2011, 127, 411–419. [Google Scholar] [CrossRef]
- Xue, H.; Zhang, B.; Tian, J.R.; Chen, M.M.; Zhang, Y.Y.; Zhang, Z.H.; Ma, Y. Comparison of the morphology, growth and development of diploid and autotetraploid ‘Hanfu’ apple trees. Sci. Hortic. 2017, 225, 277–285. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, M.; Ouwerkerk, P.B.F. Molecular and environmental factors determining grain quality in rice. Food Energy Secur. 2012, 1, 111–113. [Google Scholar] [CrossRef]
- Linden, K.J.; Callis, J. The ubiquitin system affects agronomic plant traits. J. Biol. Chem. 2020, 295, 13940–13955. [Google Scholar] [CrossRef]
- An, F.F.; Fan, J.; Li, J.; Li, Q.X.; Li, K.M.; Zhu, W.L.; Wen, F.; Carvalho, L.J.C.B.; Chen, S.B. Comparison of Leaf Proteomes of Cassava (Manihot esculenta Crantz) Cultivar NZ199 Diploid and Autotetraploid Genotypes. PLoS ONE 2014, 9, e85991. [Google Scholar] [CrossRef]
- Dong, Y.P.; Deng, M.J.; Zhao, Z.L.; Fan, G.Q. Quantitative Proteomic and Transcriptomic Study on Autotetraploid Paulownia and Its Diploid Parent Reveal Key Metabolic Processes Associated with Paulownia Autotetraploidization. Front. Plant Sci. 2016, 7, 892. [Google Scholar] [CrossRef] [Green Version]
- Huang, B.S.; Gan, L.; Chen, D.J.; Zhang, Y.C.; Zhang, Y.J.; Liu, X.L.; Chen, S.; Wei, Z.S.; Tong, L.Q.; Song, Z.J.; et al. Integration of small RNA, degradome and proteome sequencing in Oryza sativa reveals a delayed senescence network in tetraploid rice seed. PLoS ONE 2020, 15, e0242260. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Fan, G.Q.; Dong, Y.P.; Zhai, X.Q.; Deng, M.J.; Zhao, Z.L.; Liu, W.S.; Cao, Y.B. Implications of polyploidy events on the phenotype, microstructure, and proteome of Paulownia australis. PLoS ONE 2017, 12, e0172633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.T.; Sheng, Y.; Hao, Z.D.; Long, X.F.; Fu, F.F.; Liu, Y.; Tang, Z.H.; Ali, A.; Peng, Y.; Liu, Y.; et al. Transcriptome and proteome analysis suggest enhanced photosynthesis in tetraploid Liriodendron sino-americanum. Tree Physiol. 2021, 41, 1953–1971. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.X.; Yuan, J.L.; Yu, B.; Wang, X.Q.; Wang, Y.P.; Zhang, F. Leaf proteome reveals the alterations in photosynthesis and defense-related proteins between potato tetraploid cultivars and diploid wild species. J. Plant Physiol. 2022, 276, 153779. [Google Scholar] [CrossRef]
- Fan, J.; Chen, C.X.; Yu, Q.B.; Brlansky, R.H.; Li, Z.G.; Gmitter, F.G., Jr. Comparative iTRAQ proteome and transcriptome analyses of sweet orange infected by “Candidatus Liberibacter asiaticus”. Physiol. Plant 2011, 143, 235–245. [Google Scholar] [CrossRef]
- Ng, D.W.; Zhang, C.; Miller, M.; Shen, Z.; Briggs, S.P.; Chen, Z.J. Proteomic divergence in Arabidopsis autopolyploids and allopolyploids and their progenitors. Heredity 2012, 108, 419–430. [Google Scholar] [CrossRef] [Green Version]
- Lan, P.; Li, W.F.; Schmidt, W. Complementary proteome and transcriptome profiling in phosphate-deficient Arabidopsis roots reveals multiple levels of gene regulation. Mol. Cell Proteom. 2012, 11, 1156–1166. [Google Scholar] [CrossRef] [Green Version]
- Marmagne, A.; Brabant, P.; Thiellement, H.; Alix, K. Analysis of gene expression in resynthesized Brassica napus allotetraploids: Transcriptional changes do not explain differential protein regulation. New Phytol. 2010, 186, 216–227. [Google Scholar] [CrossRef]
- Wang, Q.L.; Li, Z.H. The functions of microRNAs in plants. Front. Biosci. 2007, 12, 3975–3982. [Google Scholar]
- Huang, S.L.; Zhou, J.J.; Gao, L.; Huang, S.L. Plant miR397 and its functions. Funct. Plant Biol. 2021, 48, 361–370. [Google Scholar] [CrossRef]
- Komiya, R. Biogenesis of diverse plant phasiRNAs involves an miRNA-trigger and Dicer-processing. J. Plant Res. 2017, 130, 17–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandivier, L.E.; Gregory, B.D. New insights into the plant epitranscriptome. J. Exp. Bot. 2018, 69, 4659–4665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, J.Y.; Wei, Y.; Zhao, M.M. The Reversible Methylation of m6A Is Involved in Plant Virus Infection. Biology 2022, 11, 271. [Google Scholar] [CrossRef] [PubMed]
- Shang, X.D.; Cao, Y.; Ma, L.G. Alternative Splicing in Plant Genes: A Means of Regulating the Environmental Fitness of Plants. Int. J. Mol. Sci. 2017, 18, 432. [Google Scholar] [CrossRef]






| Ploidy Level | Leaf Length (cm) | Leaf Width (cm) | Leaf Area (cm2) | Fruit Length | Fruit Width | Fruit Weight | Seed Length | Seed Width | Seed Thickness |
|---|---|---|---|---|---|---|---|---|---|
| 2× | 26.29 ± 0.73 | 19.98 ± 0.90 | 223.50 ± 8.54 | 16.75 ± 1.31 | 15.95 ± 0.95 | 2.21 ± 0.39 | 9.06 ± 0.34 | 5.44 ± 0.13 | 2.10 ± 0.08 |
| 4× | 26.01 ± 1.38 | 22.36 ± 0.25 * | 272.50 ± 6.24 * | 17.54 ± 1.56 | 17.23 ± 1.41 ** | 2.59 ± 0.59 * | 9.68 ± 0.36 ** | 6.03 ± 0.41 ** | 2.56 ± 0.23 ** |
| Spot Number | Description | Gene ID a | Biological Function | Theoretical pI /Mw (kDa) | Fold Change in Pairwise Comparison of 2x/4x | Sequence Coverage b /No. of Unique Peptides Matched c |
|---|---|---|---|---|---|---|
| 1 | Chlorophyll a/b binding protein 21 | Cla009752 | photosynthesis | 5.09/28.29 | +3.167 | 17.4/4 |
| 2 | Jasmonate-induced protein | Cla016932 | unknown | 6.39/23.90 | +5.803 | 16.0/3 |
| 3 | Low-temperature inducible | Cla010223 | unknown | 4.90/15.30 | +∝ | 40.8/5 |
| 4 | Thioredoxin m | Cla011786 | stress/transport | 9.32/19.50 | +12.103 | 35.0/7 |
| 5 | Phosphoribulokinase/uridine kinase | Cla008848 | photosynthesis | 8.19/38.85 | -∝ | 5.8/2 |
| 6 | Carbonic anhydrase | Cla017092 | carbon utilization | 6.71/36.33 | -2.650 | 14.8/4 |
| 7 | Chlorophyll a/b binding protein 8 | Cla001764 | photosynthesis | 9.38/29.31 | -2.654 | 8.4/2 |
| 8 | Cytochrome b6-f complex iron–sulfur subunit | Cla007717 | photosynthesis | 7.77/28.22 | -2.637 | 18.9/4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, Z.; Bi, Z.; Fu, D.; Feng, L.; Min, D.; Bi, C.; Huang, H. A Comparative Study of Morphology, Photosynthetic Physiology, and Proteome between Diploid and Tetraploid Watermelon (Citrullus lanatus L.). Bioengineering 2022, 9, 746. https://doi.org/10.3390/bioengineering9120746
Feng Z, Bi Z, Fu D, Feng L, Min D, Bi C, Huang H. A Comparative Study of Morphology, Photosynthetic Physiology, and Proteome between Diploid and Tetraploid Watermelon (Citrullus lanatus L.). Bioengineering. 2022; 9(12):746. https://doi.org/10.3390/bioengineering9120746
Chicago/Turabian StyleFeng, Zhanyuan, Zhubai Bi, Dugong Fu, Lihan Feng, Dangxuan Min, Chensong Bi, and He Huang. 2022. "A Comparative Study of Morphology, Photosynthetic Physiology, and Proteome between Diploid and Tetraploid Watermelon (Citrullus lanatus L.)" Bioengineering 9, no. 12: 746. https://doi.org/10.3390/bioengineering9120746
APA StyleFeng, Z., Bi, Z., Fu, D., Feng, L., Min, D., Bi, C., & Huang, H. (2022). A Comparative Study of Morphology, Photosynthetic Physiology, and Proteome between Diploid and Tetraploid Watermelon (Citrullus lanatus L.). Bioengineering, 9(12), 746. https://doi.org/10.3390/bioengineering9120746