Adaptation of Bacteria to Antineoplastic Agents Involves Persister Cells and Increases Resistance to Antibiotics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Chemicals
2.3. Determination of MIC
2.4. Tolerance to Antineoplastic Agent and Persister Formation
2.5. Biofilm Formation
2.6. Lipid Extraction and Determination of the Fatty Acid Composition
2.7. Cell Viability and Membrane Potential
2.8. Surface Tension of the Supernatant of the Cultures
2.9. Error Analysis
3. Results
3.1. Bacterial Tolerance to Antineoplastic Agents
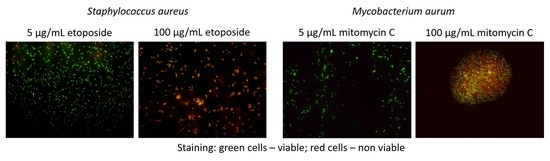
3.2. Exposure to Antineoplastic Agents Induces Persistence
3.3. Antineoplastic Induced Changes in the Cellular Envelope
3.4. Antineoplastic Induced Other Adaptation Mechanisms
3.5. Cells Adapted to Antineoplastic Become Tolerant/Resistant to Antibiotics
4. Discussion
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sylvester, R.K. Infections in patients with cancer. In ACCP’s Pharmacotherapy Self-Assessment Program, Book 10: Hematology/Oncology I and II; Schumock, G.T., Brundage, D.M., Eds.; American College of Clinical Pharmacy: Kansas City, MO, USA, 2006; pp. 147–165. [Google Scholar]
- Danai, P.A.; Moss, M.; Mannino, D.M.; Martin, G.S. The epidemiology of sepsis in patients with malignancy. CHEST 2006, 129, 1432–1440. [Google Scholar] [CrossRef] [PubMed]
- Nanayakkara, A.K.; Boucher, H.W.; Fowler Jr, V.G.; Jezek, A.; Outterson, K.; Greenberg, D.E. Antibiotic resistance in the patient with cancer: Escalating challenges and paths forward. CA A Cancer J. Clin. 2021, 71, 488–504. [Google Scholar] [CrossRef] [PubMed]
- Guinan, J.L.; McGuckin, M.; Nowell, P.C. Management of health-care-associated infections in the oncology patient. Oncology 2003, 17, 415–420. [Google Scholar] [PubMed]
- Williams, M.D.; Braun, L.A.; Cooper, L.M.; Johnston, J.; Weiss, R.V.; Qualy, R.L.; Linde-Zwirble, W. Hospitalized cancer patients with severe sepsis: Analysis of incidence, mortality, and associated costs of care. Crit. Care 2004, 8, R291–R298. [Google Scholar] [CrossRef] [Green Version]
- Kumar, G.; Ahmad, S.; Taneja, A.; Patel, J.; Guddati, A.K.; Nanchal, R.; The Milwaukee Initiative in Critical Care Outcomes Research Group of Investigators. Severe sepsis in hematopoietic stem cell transplant recipients. Crit. Care Med. 2015, 43. [Google Scholar] [CrossRef]
- Legrand, M.; Max, A.; Peigne, V.; Mariotte, E.; Canet, E.; Debrumetz, A.; Lemiale, V.; Seguin, A.; Darmon, M.; Schlemmer, B.; et al. Survival in neutropenic patients with severe sepsis or septic shock. Crit. Care Med. 2012, 40. [Google Scholar] [CrossRef]
- Bodey, G.P. Managing infections in the immunocompromised patient. Clin. Infect. Dis. 2005, 40. [Google Scholar] [CrossRef] [Green Version]
- Rolston, K.V.I. The spectrum of pulmonary infections in cancer patients. Curr. Opin. Oncol. 2001, 13. [Google Scholar] [CrossRef]
- Gudiol, C.; Aguado, J.M.; Carratalà, J. Bloodstream infections in patients with solid tumors. Virulence 2016, 7, 298–308. [Google Scholar] [CrossRef] [Green Version]
- Pilmis, B.; Le Monnier, A.; Zahar, J.-R. Gut microbiota, antibiotic therapy and antimicrobial resistance: A narrative review. Microorganisms 2020, 8, 269. [Google Scholar] [CrossRef] [Green Version]
- Armstrong, D. Life threatening infections in cancer patients. CA A Cancer J. Clin. 1973, 23, 138–150. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-L.; Chang, C.-H.; Lin, J.-W.; Wu, L.-C.; Chuang, L.-M.; Lai, M.-S. Infection, antibiotic therapy and risk of colorectal cancer: A nationwide nested case–control study in patients with Type 2 diabetes mellitus. Int. J. Cancer 2014, 135, 956–967. [Google Scholar] [CrossRef] [PubMed]
- Ashour, H.M.; El-Sharif, A. Microbial spectrum and antibiotic susceptibility profile of Gram-positive aerobic bacteria isolated from cancer patients. J. Clin. Oncol. 2007, 25, 5763–5769. [Google Scholar] [CrossRef] [PubMed]
- Zinner, S.H. Changing epidemiology of infections in patients with neutropenia and cancer: Emphasis on Gram-Positive and resistant bacteria. Clin. Infect. Dis. 1999, 29, 490–494. [Google Scholar] [CrossRef] [Green Version]
- Cruz, A.T.; Airewele, G.; Starke, J.R. Tuberculosis in pediatric oncology and bone marrow transplantation patients. Pediatric Blood Cancer 2014, 61, 1484–1485. [Google Scholar] [CrossRef]
- Wisplinghoff, H.; Seifert, H.; Wenzel, R.P.; Edmond, M.B. Current trends in the epidemiology of nosocomial bloodstream infections in patients with hematological malignancies and solid neoplasms in hospitals in the United States. Clin. Infect. Dis. 2003, 36, 1103–1110. [Google Scholar] [CrossRef] [Green Version]
- Meunier, A.; Nerich, V.; Fagnoni-Legat, C.; Richard, M.; Mazel, D.; Adotevi, O.; Bertrand, X.; Hocquet, D. Enhanced emergence of antibiotic-resistant pathogenic bacteria after in vitro induction with cancer chemotherapy drugs. J. Antimicrob. Chemother. 2019, 74, 1572–1577. [Google Scholar] [CrossRef]
- Hobson, C.A.; Bonacorsi, S.; Hocquet, D.; Baruchel, A.; Fahd, M.; Storme, T.; Tang, R.; Doit, C.; Tenaillon, O.; Birgy, A. Impact of anticancer chemotherapy on the extension of beta-lactamase spectrum: An example with KPC-type carbapenemase activity towards ceftazidime-avibactam. Sci. Rep. 2020, 10, 589. [Google Scholar] [CrossRef] [PubMed]
- Gafter-Gvili, A.; Fraser, A.; Paul, M.; Leibovici, L. Meta-Analysis: Antibiotic Prophylaxis Reduces Mortality in Neutropenic Patients. Ann. Intern. Med. 2005, 142, 979–995. [Google Scholar] [CrossRef] [PubMed]
- Gafter-Gvili, A.; Fraser, A.; Paul, M.; Vidal, L.; Lawrie Theresa, A.; van de Wetering Marianne, D.; Kremer Leontien, C.M.; Leibovici, L. Antibiotic prophylaxis for bacterial infections in afebrile neutropenic patients following chemotherapy. In Cochrane Database of Systematic Reviews; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2012. [Google Scholar]
- Gafter-Gvili, A.; Paul, M.; Fraser, A.; Leibovici, L. Effect of quinolone prophylaxis in afebrile neutropenic patients on microbial resistance: Systematic review and meta-analysis. J. Antimicrob. Chemother. 2007, 59, 5–22. [Google Scholar] [CrossRef]
- Bigger, J. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet 1944, 244, 497–500. [Google Scholar] [CrossRef]
- Kochanowski, K.; Morinishi, L.; Altschuler, S.J.; Wu, L.F. Drug persistence—From antibiotics to cancer therapies. Curr. Opin. Syst. Biol. 2018, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, F.D.A.; de Carvalho, C.C.C.R. Phenotypic modifications in Staphylococcus aureus cells exposed to high concentrations of vancomycin and teicoplanin. Front. Microbiol. 2016, 7, 13. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Persister cells, dormancy and infectious disease. Nat. Rev. Micro. 2007, 5, 48–56. [Google Scholar] [CrossRef]
- Lewis, K. Persister cells. Annu. Rev. Microbiol. 2010, 64, 357–372. [Google Scholar] [CrossRef]
- Allison, K.R.; Brynildsen, M.P.; Collins, J.J. Heterogeneous bacterial persisters and engineering approaches to eliminate them. Curr. Opin. Microbiol. 2011, 14, 593–598. [Google Scholar] [CrossRef] [Green Version]
- Windels, E.M.; Michiels, J.E.; Fauvart, M.; Wenseleers, T.; Van den Bergh, B.; Michiels, J. Bacterial persistence promotes the evolution of antibiotic resistance by increasing survival and mutation rates. ISME J. 2019, 13, 1239–1251. [Google Scholar] [CrossRef]
- CLSI supplement M100; CLSI Performance Standards for Antimicrobial Susceptibility Testing. 32nd ed. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2022.
- CLSI standard M24; CLSI Susceptibility Testing of Mycobacteria, Nocardia spp. and Other Aerobic Actinomycetes. 3rd ed. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018.
- de Carvalho, C.C.C.R.; da Fonseca, M.M.R. Assessment of three-dimensional biofilm structure using an optical microscope. Biotechniques 2007, 42, 616–620. [Google Scholar] [CrossRef] [Green Version]
- Findlay, R.H.; King, G.M.; Watling, L. Efficacy of phospholipid analysis in determining microbial biomass in sediments. Appl. Environ. Microbiol. 1989, 55, 2888–2893. [Google Scholar] [CrossRef] [Green Version]
- de Carvalho, C.C.C.R.; Fatal, V.; Alves, S.S.; da Fonseca, M.M.R. Adaptation of Rhodococcus erythropolis cells to high concentrations of toluene. Appl. Microbiol. Biotechnol. 2007, 76, 1423–1430. [Google Scholar] [CrossRef]
- de Carvalho, C.C.C.R.; Pons, M.N.; da Fonseca, M.M.R. Principal components analysis as a tool to summarise biotransformation data: Influence on cells of solvent type and phase ratio. Biocatal. Biotransformat. 2003, 21, 305–314. [Google Scholar] [CrossRef]
- Adamson, A.W. Physical Chemistry of Surfaces; Wiley: New York, NY, USA, 1976. [Google Scholar]
- Ashour, H.M.; El-Sharif, A. Species distribution and antimicrobial susceptibility of gram-negative aerobic bacteria in hospitalized cancer patients. J. Transl. Med. 2009, 7, 14. [Google Scholar] [CrossRef] [Green Version]
- Kern, W.V.; Andriof, E.; Oethinger, M.; Kern, P.; Hacker, J.; Marre, R. Emergence of fluoroquinolone-resistant Escherichia coli at a cancer center. Antimicrob. Agents Chemother. 1994, 38, 681–687. [Google Scholar] [CrossRef] [Green Version]
- Tofas, P.; Samarkos, M.; Piperaki, E.-T.; Kosmidis, C.; Triantafyllopoulou, I.-D.; Kotsopoulou, M.; Pantazatou, A.; Perlorentzou, S.; Poulli, A.; Vagia, M.; et al. Pseudomonas aeruginosa bacteraemia in patients with hematologic malignancies: Risk factors, treatment and outcome. Diagn. Microbiol. Infect. Dis. 2017, 88, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Holland, T.; Fowler, V.G., Jr.; Shelburne, S.A., 3rd. Invasive gram-positive bacterial infection in cancer patients. Clin. Infect. Dis. 2014, 59 (Suppl. 5), S331–S334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snydman, D.R.; Redelman-Sidi, G.; Sepkowitz, K.A. Rapidly growing Mycobacteria infection in patients with cancer. Clin. Infect. Dis. 2010, 51, 422–434. [Google Scholar]
- Hadied, M.O.; Bardossy, A.C.; Abreu-Lanfranco, O.; Perri, M.B.; Arshad, S.; Zervos, M.; Alangaden, G. Predictors of mortality in cancer patients with methicillin-resistant Staphylococcus aureus bloodstream infection. Open Forum. Infect. Dis. 2015, 2, 838. [Google Scholar] [CrossRef]
- Wood, T.K.; Knabel, S.J.; Kwan, B.W. Bacterial persister cell formation and dormancy. Appl. Environ. Microbiol. 2013, 79, 7116–7121. [Google Scholar] [CrossRef] [Green Version]
- Keren, I.; Kaldalu, N.; Spoering, A.; Wang, Y.; Lewis, K. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 2004, 230, 13–18. [Google Scholar] [CrossRef] [Green Version]
- de Carvalho, C.C.C.R.; Caramujo, M.J. The various roles of fatty acids. Molecules 2018, 23, 2583. [Google Scholar] [CrossRef] [Green Version]
- Ernst, R.; Ejsing, C.S.; Antonny, B. Homeoviscous adaptation and the regulation of membrane lipids. J. Mol. Biol. 2016, 428, 4776–4791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Day, A.P.; Oliver, J.D. Changes in membrane fatty acid composition during entry of Vibrio vulnificus into the viable but nonculturable state. J. Microbiol. 2004, 42, 69–73. [Google Scholar] [PubMed]
- de Carvalho, C.C.C.R.; da Cruz, A.A.R.L.; Pons, M.-N.; Pinheiro, H.M.R.V.; Cabral, J.M.S.; da Fonseca, M.M.R.; Ferreira, B.S.; Fernandes, P. Mycobacterium sp., Rhodococcus erythropolis, and Pseudomonas putida behavior in the presence of organic solvents. Microsc. Res. Tech. 2004, 64, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Lewandowski, Z.; Caldwell, D.E.; Korber, D.R.; Lappinscott, H.M. Microbial biofilms. Annu. Rev. Microbiol. 1995, 49, 711–745. [Google Scholar] [CrossRef]
- Kulka, K.; Hatfull, G.; Ojha, A.K. Growth of Mycobacterium tuberculosis biofilms. J. Vis. Exp. 2012, 60, e3820. [Google Scholar]
- EUCAST. Antimicrobial wild type distributions of microorganisms. In European Society of Clinical Microbiology and Infectious Diseases; EUCAST: Växjö, Sweden, 2022. [Google Scholar]
- Aagaard, T.; Reekie, J.; Jørgensen, M.; Roen, A.; Daugaard, G.; Specht, L.; Sengeløv, H.; Mocroft, A.; Lundgren, J.; Helleberg, M. Mortality and admission to intensive care units after febrile neutropenia in patients with cancer. Cancer Med. 2020, 9, 3033–3042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nordvig, J.; Aagaard, T.; Daugaard, G.; Brown, P.; Sengeløv, H.; Lundgren, J.; Helleberg, M. Febrile neutropenia and long-term risk of infection among patients treated with chemotherapy for malignant diseases. Open Forum. Infect. Dis. 2018, 5, ofy255. [Google Scholar] [CrossRef] [Green Version]
- Sekiguchi, M.; Takagi, Y. Effect of mitomycin C on the synthesis of bacterial and viral deoxyribonucleic acid. Biochim. Biophys. Acta 1960, 41, 434–443. [Google Scholar] [CrossRef]
- Kwan, B.W.; Chowdhury, N.; Wood, T.K. Combatting bacterial infections by killing persister cells with mitomycin C. Environ. Microbiol. 2015, 17, 4406–4414. [Google Scholar] [CrossRef]
- Chatterji, M.; Unniraman, S.; Mahadevan, S.; Nagaraja, V. Effect of different classes of inhibitors on DNA gyrase from Mycobacterium smegmatis. J. Antimicrob. Chemother. 2001, 48, 479–485. [Google Scholar] [CrossRef] [Green Version]
- Kruszewska, H.; Zareba, T.; Tyski, S. Antimicrobial activity of selected non-antibiotics--activity of methotrexate against Staphylococcus aureus strains. Acta Pol. Pharm. 2000, 27, 117–119. [Google Scholar]
- de Carvalho, C.C.C.R.; Wick, L.Y.; Heipieper, H.J. Cell wall adaptations of planktonic and biofilm Rhodococcus erythropolis cells to growth on C5 to C16 n-alkane hydrocarbons. Appl. Microbiol. Biotechnol. 2009, 82, 311–320. [Google Scholar] [CrossRef]
- de Carvalho, C.C.C.R.; Parreno-Marchante, B.; Neumann, G.; da Fonseca, M.M.R.; Heipieper, H.J. Adaptation of Rhodococcus erythropolis DCL14 to growth on n-alkanes, alcohols and terpenes. Appl. Microbiol. Biotechnol. 2005, 67, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Sikkema, J.; de Bont, J.A.; Poolman, B. Mechanisms of membrane toxicity of hydrocarbons. Microbiol. Rev. 1995, 59, 201–222. [Google Scholar] [CrossRef]
- Heipieper, H.J.; de Bont, J.A. Adaptation of Pseudomonas putida S12 to ethanol and toluene at the level of fatty acid composition of membranes. Appl. Environ. Microbiol. 1994, 60, 4440–4444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russell, N.J.; Sandercock, S.P. The regulation of bacterial membrane fluidity by modification of phospholipid fatty acyl chain length. In Membrane Fluidity: Biophysical Techniques and Cellular Regulation; Kates, M., Kuksis, A., Eds.; Humana Press: Totowa, NJ, USA, 1980; pp. 181–190. [Google Scholar]
- Denich, T.J.; Beaudette, L.A.; Lee, H.; Trevors, J.T. Effect of selected environmental and physico-chemical factors on bacterial cytoplasmic membranes. J. Microbiol. Methods 2003, 52, 149–182. [Google Scholar] [CrossRef]
- Singh, V.K.; Hattangady, D.S.; Giotis, E.S.; Singh, A.K.; Chamberlain, N.R.; Stuart, M.K.; Wilkinson, B.J. Insertional Inactivation of branched-chain alpha-keto acid dehydrogenase in Staphylococcus aureus leads to decreased branched-chain membrane fatty acid content and increased susceptibility to certain stresses. Appl. Environ. Microbiol. 2008, 74, 5882–5890. [Google Scholar] [CrossRef] [Green Version]
- Jones, T.; Yeaman, M.R.; Sakoulas, G.; Yang, S.-J.; Proctor, R.A.; Sahl, H.-G.; Schrenzel, J.; Xiong, Y.Q.; Bayer, A.S. Failures in clinical treatment of Staphylococcus aureus infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob. Agents Chemother. 2008, 52, 269–278. [Google Scholar] [CrossRef] [Green Version]
- de Carvalho, C.C.C.R.; Taglialegna, A.; Rosato, A.E. Impact of PrsA on membrane lipid composition during daptomycin-resistance-mediated β-lactam sensitization in clinical MRSA strains. J. Antimicrob. Chemother. 2022, 77, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.; de Carvalho, C.C.C.R.; Stevenson, A.; Grant, I.R.; Hallsworth, J.E. Extraordinary solute-stress tolerance contributes to the environmental tenacity of mycobacteria. Environ. Microbiol. Rep. 2015, 7, 746–764. [Google Scholar] [CrossRef] [PubMed]
- Bansal-Mutalik, R.; Nikaido, H. Mycobacterial outer membrane is a lipid bilayer and the inner membrane is unusually rich in diacyl phosphatidylinositol dimannosides. Proc. Natl. Acad. Sci. USA 2014, 111, 4958–4963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, R. Biofilms and antimicrobial resistance. Clin. Orthop. Relat. Res. 2005, 437, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Leid Jeff, G.; Shirtliff Mark, E.; Costerton, J.W.; Stoodley, P. Human leukocytes adhere to, penetrate, and respond to Staphylococcus aureus biofilms. Infect. Immun. 2002, 70, 6339–6345. [Google Scholar] [CrossRef] [Green Version]
- Mirzaei, R.; Sabokroo, N.; Ahmadyousefi, Y.; Motamedi, H.; Karampoor, S. Immunometabolism in biofilm infection: Lessons from cancer. Mol. Med. 2022, 28, 10. [Google Scholar] [CrossRef]
- Mainous, A.G., 3rd; Hueston, W.J.; Everett, C.J.; Diaz, V.A. Nasal carriage of Staphylococcus aureus and methicillin-resistant S. aureus in the United States, 2001–2002. Ann. Fam. Med. 2006, 4, 132–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boncompain, C.A.; Suárez, C.A.; Morbidoni, H.R. Staphylococcus aureus nasal carriage in health care workers: First report from a major public hospital in Argentina. Rev. Argent. Microbiol. 2017, 49, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Danelli, T.; Duarte, F.C.; Oliveira, T.A.d.; Silva, R.S.d.; Frizon Alfieri, D.; Gonçalves, G.B.; Oliveira, C.F.d.; Tavares, E.R.; Yamauchi, L.M.; Perugini, M.R.E.; et al. Nasal carriage by Staphylococcus aureus among healthcare workers and students attending a university hospital in southern Brazil: Prevalence, phenotypic, and molecular characteristics. Interdiscip. Perspect. Infect. Dis. 2020, 2020, 3808036. [Google Scholar] [CrossRef]
- Bucaneve, G.; Micozzi, A.; Menichetti, F.; Martino, P.; Dionisi, M.S.; Martinelli, G.; Allione, B.; D’Antonio, D.; Buelli, M.; Nosari, A.M.; et al. Levofloxacin to prevent bacterial infection in patients with cancer and neutropenia. N. Eng. J. Med. 2005, 353, 977–987. [Google Scholar] [CrossRef] [Green Version]
- Cullen, M.; Steven, N.; Billingham, L.; Gaunt, C.; Hastings, M.; Simmonds, P.; Stuart, N.; Rea, D.; Bower, M.; Fernando, I.; et al. Antibacterial prophylaxis after chemotherapy for solid tumors and lymphomas. N. Eng. J. Med. 2005, 353, 988–998. [Google Scholar] [CrossRef] [Green Version]
- Schlesinger, A.; Paul, M.; Gafter-Gvili, A.; Rubinovitch, B.; Leibovici, L. Infection-control interventions for cancer patients after chemotherapy: A systematic review and meta-analysis. Lancet Infect. Dis. 2009, 9, 97–107. [Google Scholar] [CrossRef]
- Dancer, S.J. How antibiotics can make us sick: The less obvious adverse effects of antimicrobial chemotherapy. Lancet Infect. Dis. 2004, 4, 611–619. [Google Scholar] [CrossRef]




| Minimum Inhibitory Concentration (MIC), μg/mL | |||||
|---|---|---|---|---|---|
| Compound | S. aureus | M. aurum | M. vaccae | P. aeruginosa | E. coli |
| Mitomycin C | 3.25 | 100 | 100 | 12.5 | 12.5 |
| Cyclophosphamide | 100 | 100 | 100 | 50 | 50 |
| Etoposide | 100 | 25 | 25 | 100 | 100 |
| Methotrexate | 100 | 12.5 | 12.5 | 6.25 | 12.5 |
| MIC (µg/mL) | S. aureus | M. aurum | M. vaccae | P. aeruginosa | E. coli |
|---|---|---|---|---|---|
| Cells not pre-exposed to antineoplastic agents | |||||
| Vancomycin | 5.2 | 2.6 | 5.2 | - | - |
| Teicoplanin | 2.6 | 10.4 | 2.6 | - | - |
| Linezolid | 2.6 | 5.2 | 2 | - | - |
| Levofloxacin | 2 | 41.7 | 2 | 5.2 | 10.3 |
| Ciprofloxacin | 2.6 | 41.7 | 2 | 20.8 | 41.7 |
| Cells pre-exposed to antineoplastic agents | |||||
| Vancomycin | 166.7 | 5.2 | 83.3 | - | - |
| Teicoplanin | 10.4 | 83.4 | 41.7 | - | - |
| Linezolid | 20.8 | 16.7 | 4.2 | - | - |
| Levofloxacin | 83.4 | 83.3 | 83.4 | 41.7 | 20.8 |
| Ciprofloxacin | 41.7 | 83.3 | 166.7 | 41.7 | 41.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Carvalho, C.C.C.R. Adaptation of Bacteria to Antineoplastic Agents Involves Persister Cells and Increases Resistance to Antibiotics. Bioengineering 2022, 9, 355. https://doi.org/10.3390/bioengineering9080355
de Carvalho CCCR. Adaptation of Bacteria to Antineoplastic Agents Involves Persister Cells and Increases Resistance to Antibiotics. Bioengineering. 2022; 9(8):355. https://doi.org/10.3390/bioengineering9080355
Chicago/Turabian Stylede Carvalho, Carla C. C. R. 2022. "Adaptation of Bacteria to Antineoplastic Agents Involves Persister Cells and Increases Resistance to Antibiotics" Bioengineering 9, no. 8: 355. https://doi.org/10.3390/bioengineering9080355
APA Stylede Carvalho, C. C. C. R. (2022). Adaptation of Bacteria to Antineoplastic Agents Involves Persister Cells and Increases Resistance to Antibiotics. Bioengineering, 9(8), 355. https://doi.org/10.3390/bioengineering9080355