Genetic Response of Common Bean to the Inoculation with Indigenous Fusarium Isolates
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fusarium spp. and Culture Collections
2.2. In Vivo Assay
2.3. RNA Extraction and Purification
2.4. cDNA Synthesis
2.5. Real Time-PCR Analysis
3. Results
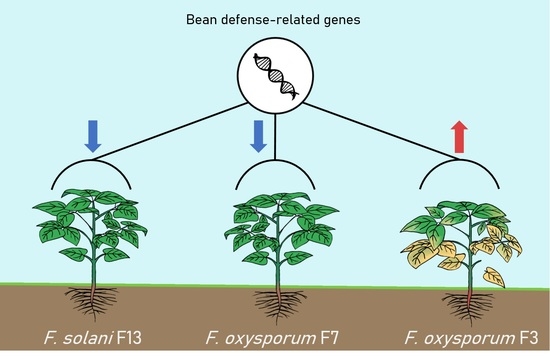
3.1. In Vivo Assay: Selection of Pathogenic and Low-Pathogenic Fusarum Isolates
3.2. Re-Isolation of the Fungal Isolates from Infected Plants
3.3. Expression of Bean Defense-Related Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 13 June 2019).
- Anuario de Estadística. Available online: https://www.mapa.gob.es/es/estadistica/temas/publicaciones/anuario-de-estadistica/2018/default.aspx (accessed on 13 June 2019).
- Mulas, D.; García-Fraile, P.; Carro, L.; Ramírez-Bahena, M.-H.; Casquero, P.; Velázquez, E.; González-Andrés, F. Distribution and efficiency of Rhizobium leguminosarum strains nodulating Phaseolus vulgaris in Northern Spanish soils: Selection of native strains that replace conventional N fertilization. Soil Biol. Biochem. 2011, 43, 2283–2293. [Google Scholar] [CrossRef]
- Valenciano, J.B.; Casquero, P.A.; Boto, J.A.; Guerra, M. Effect of sowing techniques and seed pesticide application on dry bean yield and harvest components. Field Crop. Res. 2006, 96, 2–12. [Google Scholar] [CrossRef]
- Llanos, M. Enfermedades de las judías verdes. Vida Rural 1999, 6, 42–44. [Google Scholar]
- Schwartz, H.F.; Steadman, J.R.; Hall, R.; Forster, R.L. Compendium of Bean Diseases, 2nd ed.; Schwartz, H.F., Steadman, J.R., Hall, R., Forster, R.L., Eds.; The American Phytopathological Society: St. Paul, MN, USA, 2005. [Google Scholar]
- Pathania, A.; Sharma, S.K.; Sharma, P.N. Common Bean. In Broadening the Genetic Base of Grain Legumes; Singh, M., Bisht, I., Dutta, M., Eds.; Springer: New Delhi, India, 2014; pp. 11–50. [Google Scholar]
- Kaur, R.; Kaur, J.; Singh, R.S. Nonpathogenic Fusarium as a biological control agent. Plant Pathol. 2010, 9, 88–100. [Google Scholar] [CrossRef] [Green Version]
- Producción de Semillas de Alta Calidad de Frijol Común (Phaseolus vulgaris L.); Araya-Villalobos, R.; Gutiérrez-Soto, M.V. (Eds.) Universidad de Costa Rica: Alajuela, Costa Rica, 2015; ISBN 9789968557955. [Google Scholar]
- De Ron, A.M.; Kalavacharla, V.; Álvarez-García, S.; Casquero, P.A.; Carro-Huerga, G.; Gutiérrez, S.; Lorenzana, A.; Mayo-Prieto, S.; Rodríguez-González, A.; Suárez-Villanueva, V.; et al. Common bean genetics, breeding, and genomics for adaptation to changing to new agri-environmental conditions. In Genomic Designing of Climate-Smart Pulse Crops; Springer International Publishing: Cham, Switzerland, 2019; pp. 1–106. [Google Scholar]
- Aimé, S.; Cordier, C.; Alabouvette, C.; Olivain, C. Comparative analysis of PR gene expression in tomato inoculated with virulent Fusarium oxysporum f. sp. lycopersici and the biocontrol strain F. Oxysporum Fo47. Physiol. Mol. Plant Pathol. 2008, 73, 9–15. [Google Scholar]
- Barros-Ríos, J.; Malvar, R.A.; Santiago, R. Función de la pared celular del maíz (Zea mays L.) como mecanismo de defensa frente a la plaga del taladro (Ostrinia nubilalis Hüb y Sesamia nonagrioides Lef.). Rev. Educ. Bioquímica 2011, 30, 132–142. [Google Scholar]
- Swarupa, V.; Ravishankar, K.V.; Rekha, A. Plant defense response against Fusarium oxysporum and strategies to develop tolerant genotypes in banana. Planta 2014, 239, 735–751. [Google Scholar] [CrossRef] [Green Version]
- Conrath, U.; Beckers, G.J.M.; Flors, V.; García-Agustín, P.; Jakab, G.; Mauch, F.; Newman, M.-A.; Pieterse, C.M.J.; Poinssot, B.; Pozo, M.J.; et al. Priming: Getting ready for battle. Mol. Plant Microbe Interact. 2006, 19, 1062–1071. [Google Scholar] [CrossRef] [Green Version]
- Olivain, C.; Alabouvette, C. Process of tomato root colonization by a pathogenic strain of Fusarium oxysporum f. sp. lycopersici in comparison with a non-pathogenic strain. New Phytol. 1999, 141, 497–510. [Google Scholar] [CrossRef]
- Hermosa, R.; Belén Rubio, M.; Cardoza, R.E.; Nicolás, C.; Monte, E.; Gutiérrez, S. The contribution of Trichoderma to balancing the costs of plant growth and defense. Int. Microbiol. 2013, 16, 69–80. [Google Scholar]
- Mukherjee, P.K.; Horwitz, B.A.; Herrera-Estrella, A.; Schmoll, M.; Kenerley, C.M. Trichoderma research in the genome era. Annu. Rev. Phytopathol. 2013, 51, 105–129. [Google Scholar] [CrossRef] [PubMed]
- Sneh, B. Use of non-pathogenic or hypovirulent fungal strains to protect plants against closely related fungal pathogens. Biotechnol. Adv. 1998, 16, 1–32. [Google Scholar] [CrossRef]
- Benhamou, N.; Garand, C.; Goulet, A. Ability of nonpathogenic Fusarium oxysporum strain Fo47 to induce resistance against Pythium ultimum infection in cucumber. Appl. Environ. Microbiol. 2002, 68, 4044–4060. [Google Scholar] [CrossRef] [Green Version]
- Paparu, P.; Dubois, T.; Coyne, D.; Viljoen, A. Defense-related gene expression in susceptible and tolerant bananas (Musa spp.) following inoculation with non-pathogenic Fusarium oxysporum endophytes and challenge with Radopholus similis. Physiol. Mol. Plant Pathol. 2007, 71, 149–157. [Google Scholar] [CrossRef]
- Ongena, M.; Jourdan, E.; Schäfer, M.; Kech, C.; Budzikiewicz, H.; Luxen, A.; Thonart, P. Isolation of an N-alkylated benzylamine derivative from Pseudomonas putida BTP1 as elicitor of induced systemic resistance in bean. Mol. Plant Microbe Interact. 2005, 18, 562–569. [Google Scholar] [CrossRef] [Green Version]
- Clemente, M. Evaluation of Isolates of Fusarium in Three Bean Landraces (Phaseolus vulgaris L.) of León (Spain), Trabajo fin de Carrera; Universidad de León: León, France, 2007. [Google Scholar]
- Rigaud, J.; Puppo, A. Indole-3-acetic acid catabolism by soybean bacteroids. J. Gen. Microbiol. 1975, 88, 223–228. [Google Scholar] [CrossRef] [Green Version]
- Mayo, S.; Cominelli, E.; Sparvoli, F.; González-López, O.; Rodríguez-González, A.; Gutiérrez, S.; Casquero, P.A. Development of a qPCR strategy to select bean genes involved in plant defense response and regulated by the Trichoderma velutinum—Rhizoctonia solani interaction. Front. Plant Sci. 2016, 7, 1109. [Google Scholar] [CrossRef]
- Mayo, S.; Gutiérrez, S.; Malmierca, M.G.; Lorenzana, A.; Campelo, M.P.; Hermosa, R.; Casquero, P.A. Influence of Rhizoctonia solani and Trichoderma spp. in growth of bean (Phaseolus vulgaris L.) and in the induction of plant defense-related genes. Front. Plant Sci. 2015, 6, 685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reid, K.E.; Olsson, N.; Schlosser, J.; Peng, F.; Lund, S.T. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 2006, 6, 27. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Borges, A.; Tsai, S.M.; Caldas, D.G.G. Validation of reference genes for RT-qPCR normalization in common bean during biotic and abiotic stresses. Plant Cell Rep. 2012, 31, 827–838. [Google Scholar] [CrossRef] [PubMed]
- Ting, A.S.Y.; Meon, S.; Kadir, J.; Radu, S.; Singh, G. Endophytic microorganisms as potential growth promoters of banana. BioControl 2008, 53, 541–553. [Google Scholar] [CrossRef]
- Bakshi, M.; Oelmüller, R. WRKY transcription factors. Plant Signal. Behav. 2014, 9, e27700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, S.P.; Somssich, I.E. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birkenbihl, R.P.; Diezel, C.; Somssich, I.E. Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 2012, 159, 266–285. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Z.; Qamar, S.A.; Chen, Z.; Mengiste, T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006, 48, 592–605. [Google Scholar] [CrossRef]
- Berrocal-Lobo, M.; Molina, A. Ethylene Response Factor 1 mediates Arabidopsis resistance to the soilborne fungus Fusarium oxysporum. Mol. Plant Microbe Interact. 2004, 17, 763–770. [Google Scholar] [CrossRef] [Green Version]
- Boller, T. Ethylene and the regulation of antifungal hydrolases in plants. Oxf. Surv. Plant Mol. Cell Biol. 1989, 5, 145–175. [Google Scholar]
- Wang, Y.; Kang, Y.; Ma, C.; Miao, R.; Wu, C.; Long, Y.; Ge, T.; Wu, Z.; Hou, X.; Zhang, J.; et al. CNGC2 is a Ca2+ influx channel that prevents accumulation of apoplastic Ca2+ in the leaf. Plant Physiol. 2017, 173, 1342–1354. [Google Scholar] [CrossRef] [Green Version]
- Mauch, F.; Dudler, R. Differential induction of distinct glutathione-S-transferases of wheat by xenobiotics and by pathogen attack. Plant Physiol. 1993, 102, 1193–1201. [Google Scholar] [CrossRef] [Green Version]
- Moons, A. Regulatory and functional interactions of plant growth regulators and plant glutathione S-transferases (GSTs). Vitam. Horm. 2005, 72, 155–202. [Google Scholar]
- Dixon, D.P.; Lapthorn, A.; Edwards, R. Plant glutathione transferases. Genome Biol. 2002, 3. reviews3004.1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gullner, G.; Komives, T.; Király, L.; Schröder, P. Glutathione S-transferase enzymes in plant-pathogen interactions. Front. Plant Sci. 2018, 9, 1836. [Google Scholar] [CrossRef] [Green Version]
- Narasimhan, M.L.; Bressan, R.A.; D’Urzo, M.P.; Jenks, M.A.; Mengiste, T. Chapter 11 Unexpected Turns and Twists in Structure/Function of PR-Proteins that Connect Energy Metabolism and Immunity. In Advances in Botanical Research; Academic Press: Cambridge, MA, USA, 2009; Volume 51, pp. 439–489. [Google Scholar]
- Chowdhury, S.; Basu, A.; Kundu, S. Cloning, characterization, and bacterial over-expression of an osmotin-like protein gene from Solanum nigrum L. with antifungal activity against three necrotrophic fungi. Mol. Biotechnol. 2015, 57, 371–381. [Google Scholar] [CrossRef]
- Wen, K.; Seguin, P.; St.-Arnaud, M.; Jabaji-Hare, S. Real-Time quantitative RT-PCR of defense-associated gene transcripts of Rhizoctonia solani-infected bean seedlings in response to inoculation with a nonpathogenic binucleate Rhizoctonia isolate. Phytopathology 2005, 95, 345–353. [Google Scholar] [CrossRef]
- Alabouvette, C.; Couteaudier, Y. Biological control of Fusarium wilts with nonpathogenic Fusaria. In Biological Control of Plant Diseases; Tjamos, E.C., Papavizas, G.C., Cook, R.J., Eds.; Springer: Boston, MA, USA, 1992; pp. 415–426. [Google Scholar]
- He, C.Y.; Hsiang, T.; Wolyn, D.J. Induction of systemic disease resistance and pathogen defence responses in Asparagus officinalis inoculated with nonpathogenic strains of Fusarium oxysporum. Plant Pathol. 2002, 51, 225–230. [Google Scholar] [CrossRef] [Green Version]




| Code | Identified As 1 | % Identity | Bean Landrace |
|---|---|---|---|
| F1 | F. oxysporum | >99% | Canela |
| F2 | F. oxysporum | >99% | Canela |
| F3 | F. oxysporum | >99% | Canela |
| F4 | F. oxysporum | >99% | Pinta |
| F5 | F. oxysporum | >99% | Canela |
| F6 | F. oxysporum | >99% | Canela |
| F7 | F. oxysporum | >99% | Pinta |
| F9 | F. oxysporum | >99% | Riñón menudo |
| F10 | F. oxysporum | >99% | Riñón menudo |
| F11 | F. oxysporum | >99% | Riñón |
| F12 | F. oxysporum | >99% | Pinta |
| F13 | F. solani | >99% | Pinta |
| Gene | Functional Annotation | JGI Phytozome | Forward/Reverse |
|---|---|---|---|
| Act112 | Actin-11 | Phvul.008G011000 | TGCATACGTTGGTGATGAGG AGCCTTGGGGTTAAGAGGAG |
| Amintransf22 | Aminotransferase 2 | Phvul.006G029100 | TTCTTCCTTTTCTGCTCTTTCAA AGATGACAAGATGCAATGATTTTT |
| Ukn12 | Unknown | Phvul.011G023200 | ATTCCCATCATGCAGCAAAG AGATCCCTCCAGGTCAATCC |
| CH5b1 | Endochitinase precursor | Phvul.009G116500 | CAGCCAAAGGCTTCTACACC TTGTTTCGTGAGACGTTTGC |
| CNGC22 | Cyclic nucleotide-gated ion channel 2 | Phvul.008G036200 | ATTCAATTTGCTTGGAGACGTT ACAGTTTTATTGAAGGCCAGGA |
| ERF12 | Ethylene-Responsive Transcription factor 1 | Phvul.007G127800 | CGCTCTCAAGAGGAAACACTCC TGAATCAGAAGGAGGAGGGAAT |
| ERF52 | Ethylene-Responsive Transcription factor 5 | Phvul.002G055700 | GGCTCCAAGTGGATTGAGAAC TCAGAATCAGATAACTACAAAGCACAA |
| GSTa2 | 2,4-D inducible glutathione S-transferase | Phvul.002G241400 | AGGGAGTCACACTGGCTATGTT ATGTGCCATTTGCATTTTAGTG |
| hGS2 | Homoglutathione synthetase | Phvul.006G094500 | GTGGCTATATGGTGCGTACAAA GAAACAAGAATGCATCTCCTCA |
| HPL2 | Hydroperoxide lyase | Phvul.005G116800 | TCAAGGCTACATTTGTATTTCCA TGGTGCACATTTCTTAGTAGCAA |
| IPER2 | Peroxidase precursors | Phvul.009G215000 | GGCAAGCATTATATGGTTGAAA GATGGCAACATCCATCACTTTA |
| Lox22 | Lipoxygenase 2 | Phvul.005G156700 | ATGCAAGGCTAAAGAGATCCAA ATGGTGACAGGAGCTAAACACA |
| Lox72 | Lipoxygenase 7 | Phvul.005G156900 | GAAGGCTTGACTTTCAGAGGAA AACACACGAGAAGATTCAACCA |
| OSM342 | Osmotin-like protein | Phvul.002G155500 | GAACGGAGGGTGTCACAAAATC CGTAGTGGGTCCACAAGTTCCT |
| PAL12 | Phenylalanine ammonia-lyase | Phvul.001G177800 | TGAGAGAGGAGTTGGGCACT TTCCACTCTCCAAGGCATTC |
| PPO2 | Polyphenol oxidase | Phvul.008G073200 | GAAGACGATGATTTGCTGGTTA AAGAAACATTTTCCTTTGTGAAA |
| PR11 | Pathogenesis-related 1 | Phvul.003G109100 | TGGTCCTAACGGAGGATCAC TGGCTTTTCCAGCTTTGAGT |
| PR21 | Β-1,3 endoglucanase | Phvul.003G109200 | GTGAAGGACGCCGATAACAT ACTGAGTTTGGGGTCGATTG |
| PR32 | Basic Endochitinase B | Phvul.009G116600 | TGGAGTTGGTTATGGCAACAA ATTCTGATGGGATGGCAGTGT |
| PR41 | Pathogenesis-related 4 | Phvul.006G102300 | CGCAGTGAGTGCATATTGCT TGTTTGTCACCCTCAAGCAC |
| PR16a2 | Germin-like protein 8 | Phvul.010G129900 | GGCAGTCTCATGGTTATGGTTT GCATGCTCAAGTCTCAACACAT |
| WRKY332 | WRKY transcription factors | Phvul.008G090300 | TTTCACAGGACAGGTTCCAGC CCTTTGACAGAAATGACTGAAGGA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Porteous-Álvarez, A.J.; Mayo-Prieto, S.; Álvarez-García, S.; Reinoso, B.; Casquero, P.A. Genetic Response of Common Bean to the Inoculation with Indigenous Fusarium Isolates. J. Fungi 2020, 6, 228. https://doi.org/10.3390/jof6040228
Porteous-Álvarez AJ, Mayo-Prieto S, Álvarez-García S, Reinoso B, Casquero PA. Genetic Response of Common Bean to the Inoculation with Indigenous Fusarium Isolates. Journal of Fungi. 2020; 6(4):228. https://doi.org/10.3390/jof6040228
Chicago/Turabian StylePorteous-Álvarez, Alejandra J., Sara Mayo-Prieto, Samuel Álvarez-García, Bonifacio Reinoso, and Pedro A. Casquero. 2020. "Genetic Response of Common Bean to the Inoculation with Indigenous Fusarium Isolates" Journal of Fungi 6, no. 4: 228. https://doi.org/10.3390/jof6040228
APA StylePorteous-Álvarez, A. J., Mayo-Prieto, S., Álvarez-García, S., Reinoso, B., & Casquero, P. A. (2020). Genetic Response of Common Bean to the Inoculation with Indigenous Fusarium Isolates. Journal of Fungi, 6(4), 228. https://doi.org/10.3390/jof6040228