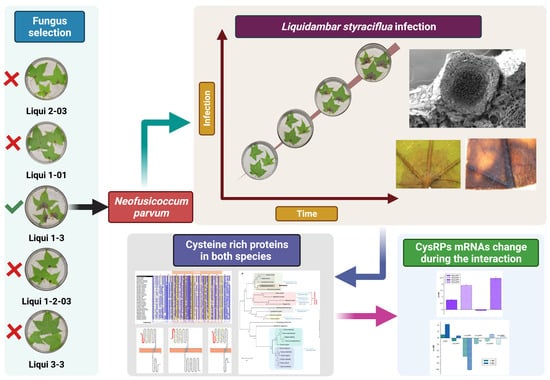
Insights of the Neofusicoccum parvum–Liquidambar styraciflua Interaction and Identification of New Cysteine-Rich Proteins in Both Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Isolation of Fungi from L. styraciflua Leaves
2.3. DNA Extraction and Molecular Identification of the Liqui 1–3–Isolate by PCR
2.4. Pathogenicity Assay in Arabidopsis Thaliana Seedlings
2.5. Pathogenicity Assay in L. styraciflua Leaves and Stems
2.6. DAB Staining
2.7. SEM
2.8. CysRP Identification
2.9. Phylogenetic Trees
2.10. RNA Extraction and Gene Expression
3. Results
3.1. Identification of N. parvum as a Pathogen of L. styraciflua
3.2. Establishment of the L. styraciflua–N. parvum Pathosystem
3.3. Identification and Description of CysRPs in L. styraciflua and N. parvum
3.4. CysRP Phylogenetic Analyses
3.5. Expression of CysRP mRNAs of L. styraciflua and N. parvum during Early Stages of the Infection Process
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Slippers, B.; Wingfield, M.J. Botryosphaeriaceae as endophytes and latent pathogens of woody plants: Diversity, ecology and impact. Fungal Biol. Rev. 2007, 21, 90–106. [Google Scholar] [CrossRef]
- Sakalidis, M.L.; Slippers, B.; Wingfield, B.D.; Hardy, G.E.S.J.; Burgess, T.I. The challenge of understanding the origin, pathways and extent of fungal invasions: Global populations of the Neofusicoccum parvum-N. ribisspecies complex. Divers. Distrib. 2013, 19, 873–883. [Google Scholar] [CrossRef]
- Lorenzini, M.; Cappello, M.S.; Zapparoli, G. Isolation of Neofusicoccum parvum from withered grapes: Strain characterization, pathogenicity and its detrimental effects on passito wine aroma. J. Appl. Microbiol. 2015, 119, 1335–1344. [Google Scholar] [CrossRef] [Green Version]
- Carrillo, J.; Eskalen, A.; Rooney-Latham, S.; Scheck, H.J. First Report of Neofusicoccum nonquaesitum Causing Branch Canker and Dieback of Avocado in California. Plant Dis. 2016, 100, 1778. [Google Scholar] [CrossRef]
- Molina-Gayosso, E.; Silva-Rojas, H.V.; García-Morales, S.; Avila-Quezada, G. First Report of Black Spots on Avocado Fruit Caused by Neofusicoccum parvum in Mexico. Plant Dis. 2012, 96, 287. [Google Scholar] [CrossRef] [PubMed]
- Valencia, A.L.; Gil, P.M.; Latorre, B.A.; Rosales, I.M. Characterization and Pathogenicity of Botryosphaeriaceae Species Obtained from Avocado Trees with Branch Canker and Dieback and from Avocado Fruit with Stem End Rot in Chile. Plant Dis. 2019, 103, 996–1005. [Google Scholar] [CrossRef]
- Espinoza, J.G.; Briceño, E.X.; Chávez, E.R.; Úrbez-Torres, J.R.; Latorre, B.A. Neofusicoccum spp. Associated with Stem Canker and Dieback of Blueberry in Chile. Plant Dis. 2009, 93, 1187–1194. [Google Scholar] [CrossRef] [Green Version]
- Palavouzis, S.; Tzamos, S.; Paplomatas, E.; Thomidis, T. First report of Neofusicoccum parvum causing shoot blight of pomegranate in Northern Greece. New Dis. Rep. 2015, 32, 10. [Google Scholar] [CrossRef] [Green Version]
- Song, Z.X.; Liao, J.; Luo, H.; Zhang, F.; Sun, Z.X.; Liu, Q.K.; Deng, J.X. First Report of Neofusicoccum parvum Associated with Shoot Blight on Peaches in China. Plant Dis. 2019, 103, 1429. [Google Scholar] [CrossRef]
- Cheon, W.; Kim, Y.S.; Lee, S.G.; Jeon, Y.H.; Chun, I.-J. First Report of Branch Dieback of Walnut Caused by Neofusicoccum parvum in Korea. Plant Dis. 2013, 97, 1114. [Google Scholar] [CrossRef]
- Aćimović, S.G.; Rooney-Latham, S.; Albu, S.; Grosman, D.M.; Doccola, J.J. Characterization and Pathogenicity of Botryosphaeriaceae Fungi Associated with Declining Urban Stands of Coast Redwood in California. Plant Dis. 2018, 102, 1950–1957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golzar, H.; Burgess, T.I. Neofusicoccum parvum, a causal agent associated with cankers and decline of Norfolk Island pine in Australia. Australas. Plant Pathol. 2011, 40, 484–489. [Google Scholar] [CrossRef]
- Iturritxa, E.; Slippers, B.; Mesanza, N.; Wingfield, M.J. First report of Neofusicoccum parvum causing canker and die-back of Eucalyptus in Spain. Australas. Plant Dis. Notes 2011, 6, 57–59. [Google Scholar] [CrossRef] [Green Version]
- Lopes, A. Diversity and phylogeny of Neofusicoccum species occurring in forest and urban environments in Portugal. Mycosphere 2016, 7, 906–920. [Google Scholar] [CrossRef]
- Mirhosseini, H.; Babaeizad, V.; Rahimlou, S. Neofusicoccum parvum, agent of leaf spot on the new host Ginkgo biloba in Iran. New Dis. Rep. 2014, 30, 12. [Google Scholar] [CrossRef] [Green Version]
- Blanco-Ulate, B.; Rolshausen, P.; Cantu, D. Draft Genome Sequence of Neofusicoccum parvum Isolate UCR-NP2, a Fungal Vascular Pathogen Associated with Grapevine Cankers. Genome Announc. 2013, 1, e00339-13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abou-Mansour, E.; Débieux, J.-L.; Ramírez-Suero, M.; Bénard-Gellon, M.; Magnin-Robert, M.; Spagnolo, A.; Chong, J.; Farine, S.; Bertsch, C.; L’Haridon, F.; et al. Phytotoxic metabolites from Neofusicoccum parvum, a pathogen of Botryosphaeria dieback of grapevine. Phytochemistry 2015, 115, 207–215. [Google Scholar] [CrossRef]
- Trotel-Aziz, P.; Mansour, E.A.; Courteaux, B.; Rabenoelina, F.; Clément, C.; Fontaine, F.; Aziz, A. Bacillus subtilis PTA-271 Counteracts Botryosphaeria Dieback in Grapevine, Triggering Immune Responses and Detoxification of Fungal Phytotoxins. Front. Plant Sci. 2019, 10, 25. [Google Scholar] [CrossRef] [PubMed]
- Pour, F.N.; Cobos, R.; Coque, J.J.R.; Serôdio, J.; Alves, A.; Félix, C.; Ferreira, V.; Esteves, A.C.; Duarte, A.S. Toxicity of Recombinant Necrosis and Ethylene-Inducing Proteins (NLPs) from Neofusicoccum parvum. Toxins 2020, 12, 235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehl, J.W.; Slippers, B.; Roux, J.; Wingfield, M.J. Overlap of latent pathogens in the Botryosphaeriaceae on a native and agricultural host. Fungal Biol. 2017, 121, 405–419. [Google Scholar] [CrossRef] [Green Version]
- Adams, J.; Lingbeck, J.; Crandall, P.; Martin, E.; O’Bryan, C. Sweetgum: A new look. iForest Biogeosci. For. 2015, 8, 719–727. [Google Scholar] [CrossRef]
- Gao, L.; Li, Y.; Xu, Y.; Hulcr, J.; Cognato, A.I.; Wang, J.-G.; Ju, R.-T. Acanthotomicus sp. (Coleoptera: Curculionidae: Scolytinae), a New Destructive Insect Pest of North American Sweetgum Liquidambar styraciflua in China. J. Econ. Entomol. 2017, 110, 1592–1595. [Google Scholar] [CrossRef]
- Kline, K.L.; Coleman, M.D. Woody energy crops in the southeastern United States: Two centuries of practitioner experience. Biomass Bioenergy 2010, 34, 1655–1666. [Google Scholar] [CrossRef] [Green Version]
- Pedraza, R.; Williams-Linera, G. Evaluation of native tree species for the rehabilitation of deforested areas in a Mexican cloud forest. New For. 2003, 26, 83–99. [Google Scholar] [CrossRef]
- Lu, S.; Edwards, M.C. Genome-Wide Analysis of Small Secreted Cysteine-Rich Proteins Identifies Candidate Effector Proteins Potentially Involved in Fusarium graminearum−Wheat Interactions. Phytopathology 2016, 106, 166–176. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.-R.; Li, Y.-P.; Li, Q.-Y.; Xing, Y.-P.; Liu, B.-B.; Tong, Y.-H.; Xu, J.-Y.; And, Y.T. SCR96, a small cysteine-rich secretory protein of P hytophthora cactorum, can trigger cell death in the Solanaceae and is important for pathogenicity and oxidative stress tolerance. Mol. Plant Pathol. 2015, 17, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Lyu, X.; Shen, C.; Fu, Y.; Xie, J.; Jiang, D.; Li, G.; Cheng, J. A Small Secreted Virulence-Related Protein is Essential for the Necrotrophic Interactions of Sclerotinia sclerotiorum with Its Host Plants. PLoS Pathog. 2016, 12, e1005435. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Tian, L.; Zhang, D.; Song, J.; Song, S.; Yin, C.; Zhou, L.; Liu, Y.; Wang, B.; Kong, Z.; et al. Functional analyses of small secreted cysteine-rich proteins identified candidate effectors in Verticillium dahliae. Mol. Plant Pathol. 2020, 21, 667–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Odintsova, T.I.; Slezina, M.P.; Istomina, E.A. Defensins of Grasses: A Systematic Review. Biomolecules 2020, 10, 1029. [Google Scholar] [CrossRef] [PubMed]
- Slazak, B.; Kapusta, M.; Strömstedt, A.; Słomka, A.; Krychowiak-Masnicka, M.; Shariatgorji, R.; Andrén, P.E.; Bohdanowicz, J.; Kuta, E.; Göransson, U. How Does the Sweet Violet (Viola odorata L.) Fight Pathogens and Pests—Cyclotides as a Comprehensive Plant Host Defense System. Front. Plant Sci. 2018, 9, 1296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, T.; Han, M.; Cao, D.; Xu, M. Molecular and Biological Properties of Snakins: The Foremost Cysteine-Rich Plant Host Defense Peptides. J. Fungi 2020, 6, 220. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.H.; Tan, W.L.; Serra, A.; Xiao, T.; Sze, S.K.; Yang, D.; Tam, J.P. Ginkgotides: Proline-Rich Hevein-Like Peptides from Gymnosperm Ginkgo biloba. Front. Plant Sci. 2016, 7, 1639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tapia-Tussell, R.; Lappe, P.; Ulloa, M.; Quijano-Ramayo, A.; Cáceres-Farfán, M.; Larqué-Saavedra, A.; Perez-Brito, D. A rapid and simple method for DNA extraction from yeasts and fungi isolated from Agave fourcroydes. Mol. Biotechnol. 2006, 33, 67–70. [Google Scholar] [CrossRef]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [Green Version]
- Sakalidis, M.; Ray, J.D.; Lanoiselet, V.; Hardy, G.; Burgess, T.I. Pathogenic Botryosphaeriaceae associated with Mangifera indica in the Kimberley Region of Western Australia. Eur. J. Plant Pathol. 2011, 130, 379–391. [Google Scholar] [CrossRef] [Green Version]
- Slippers, B.; Crous, P.W.; Denman, S.; Coutinho, T.A.; Wingfield, B.D.; Wingfield, M.J. Combined Multiple Gene Genealogies and Phenotypic Characters Differentiate Several Species Previously Identified as Botryosphaeria dothidea. Mycologia 2004, 96, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daudi, A.; O’Brien, J.A. Detection of Hydrogen Peroxide by DAB Staining in Arabidopsis Leaves. Bio-Protoc. 2012, 2, e263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bozzola, J.J.; Russell, L.D. Electron Microscopy: Principles and Techniques for Biologists; Jones and Bartlett series in biology; Jones and Bartlett: Burlington, MA, USA, 1999; ISBN 9780763701925. [Google Scholar]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; von Heijne, G. Predicting Subcellular Localization of Proteins Based on their N-terminal Amino Acid Sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef] [Green Version]
- Omasits, U.; Ahrens, C.; Müller, S.; Wollscheid, B. Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 2014, 30, 884–886. [Google Scholar] [CrossRef] [Green Version]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Armenteros, J.J.A.; Sønderby, C.K.; Sønderby, S.K.; Nielsen, H.; Winther, O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef]
- Kulmanov, M.; Hoehndorf, R. DeepGOPlus: Improved protein function prediction from sequence. Bioinformatics 2019, 36, 422–429. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: New York, NY, USA, 2005; pp. 571–607. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Notredame, C.; Higgins, D.; Heringa, J. T-coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef] [Green Version]
- Thornton, B.; Basu, C. Real-time PCR (qPCR) primer design using free online software. Biochem. Mol. Biol. Educ. 2011, 39, 145–154. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef] [Green Version]
- Albuquerque, G.M.R.; Fonseca, F.C.A.; Boiteux, L.S.; Borges, R.C.F.; Miller, R.N.G.; Lopes, C.A.; Souza, E.B.; Fonseca, M.E.N. Stability analysis of reference genes for RT-qPCR assays involving compatible and incompatible Ralstonia solanacearum-tomato ‘Hawaii 7996’ interactions. Sci. Rep. 2021, 11, 18719. [Google Scholar] [CrossRef] [PubMed]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.; Scheible, W.-R. Genome-Wide Identification and Testing of Superior Reference Genes for Transcript Normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freeman, S.; Sharon, M.; Maymon, M.; Mendel, Z.; Protasov, A.; Aoki, T.; Eskalen, A.; O’Donnell, K. Fusarium euwallaceae sp. nov.—A symbiotic fungus of Euwallacea sp., an invasive ambrosia beetle in Israel and California. Mycologia 2013, 105, 1595–1606. [Google Scholar] [CrossRef] [Green Version]
- Na, F.; Carrillo, J.; Mayorquin, J.S.; Ndinga-Muniania, C.; Stajich, J.E.; Stouthamer, R.; Huang, Y.-T.; Lin, Y.-T.; Chen, C.-Y.; Eskalen, A. Two Novel Fungal Symbionts Fusarium kuroshium sp. nov. and Graphium kuroshium sp. nov. of Kuroshio Shot Hole Borer (Euwallacea sp. nr. fornicatus) Cause Fusarium Dieback on Woody Host Species in California. Plant Dis. 2018, 102, 1154–1164. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.-N.; Wu, Q.-Y.; Zhang, G.-Z.; Zhu, Y.-Y.; Murphy, R.W.; Liu, Z.; Zou, C.-G. Systematic analyses reveal uniqueness and origin of the CFEM domain in fungi. Sci. Rep. 2015, 5, 13032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cram, M.; Coyle, D.; Spaine, P.; Lumpkin, S.; Coleman, M. Fertilization and Irrigation Effect on Botryosphaeriaceae Canker Development in Intensively Managed Sweetgum (Liquidambar Styraciflua). Tree Plant. Notes 2018, 61, 26–34. [Google Scholar]
- Hepting, G.H. Diseases of Forest and Shade Trees of the United States; U.S. Department of Agriculture; Forest Service: Washington, DC, USA, 1971; p. 658. [Google Scholar]
- Amponsah, N.; Jones, E.; Ridgway, H.; Jaspers, M.V. Microscopy of some interactions between Botryosphaeriaceae species and grapevine tissues. Australas. Plant Pathol. 2012, 41, 665–673. [Google Scholar] [CrossRef]
- Pedraza, J.M.T.; Aguilera, A.M.; Díaz, C.N.; Ortiz, D.T.; Ponce, G.V.; Monter, Á.V.; López, J.H. Identification, Pathogenicity, and Histopathology of Lasiodiplodia Theobromae on Mamey Sapote Grafts in Guerrero, México. Agrociencia 2012, 46, 147–161. [Google Scholar]
- Van Loon, L.; van Strien, E. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 1999, 55, 85–97. [Google Scholar] [CrossRef]
- De Lucca, A.J.; Cleveland, T.E.; Wedge, D.E. Plant-derived antifungal proteins and peptides. Can. J. Microbiol. 2005, 51, 1001–1014. [Google Scholar] [CrossRef] [Green Version]
- Molina, A.; Segura, A.; García-Olmedo, F. Lipid transfer proteins (nsLTPs) from barley and maize leaves are potent inhibitors of bacterial and fungal plant pathogens. FEBS Lett. 1993, 316, 119–122. [Google Scholar] [CrossRef] [Green Version]
- Jia, Z.; Gou, J.; Sun, Y.; Yuan, L.; Tang, Q.; Yang, X.; Pei, Y.; Luo, K. Enhanced resistance to fungal pathogens in transgenic Populus tomentosa Carr. by overexpression of an nsLTP-like antimicrobial protein gene from motherwort (Leonurus japonicus). Tree Physiol. 2010, 30, 1599–1605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Wang, X. One new kind of phytohormonal signaling integrator: Up-and-coming GASA family genes. Plant Signal. Behav. 2017, 12, e1226453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nahirñak, V.; Almasia, N.I.; Hopp, H.E.; Vazquez-Rovere, C. Snakin/GASA proteins: Involvement in Hormone Crosstalk and Redox Homeostasis. Plant Signal. Behav. 2012, 7, 1004–1008. [Google Scholar] [CrossRef] [Green Version]
- Bindschedler, L.V.; Whitelegge, J.P.; Millar, D.J.; Bolwell, G.P. A two component chitin-binding protein from French bean—association of a proline-rich protein with a cysteine-rich polypeptide. FEBS Lett. 2006, 580, 1541–1546. [Google Scholar] [CrossRef] [Green Version]
- Massonnet, M.; Morales-Cruz, A.; Figueroa-Balderas, R.; Lawrence, D.P.; Baumgartner, K.; Cantu, D. Condition-dependent co-regulation of genomic clusters of virulence factors in the grapevine trunk pathogen Neofusicoccum parvum. Mol. Plant Pathol. 2018, 19, 21–34. [Google Scholar] [CrossRef] [Green Version]
- Rep, M. Small proteins of plant-pathogenic fungi secreted during host colonization. FEMS Microbiol. Lett. 2005, 253, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, R.D.; Kelkar, H.S.; Dean, A.R. An eight-cysteine-containing CFEM domain unique to a group of fungal membrane proteins. Trends Biochem. Sci. 2003, 28, 118–121. [Google Scholar] [CrossRef]
- Fass, D. Disulfide Bonding in Protein Biophysics. Annu. Rev. Biophys. 2012, 41, 63–79. [Google Scholar] [CrossRef] [PubMed]
- Fass, D.; Thorpe, C. Chemistry and Enzymology of Disulfide Cross-Linking in Proteins. Chem. Rev. 2018, 118, 1169–1198. [Google Scholar] [CrossRef]
- Zhu, W.; Wei, W.; Wu, Y.; Zhou, Y.; Peng, F.; Zhang, S.; Chen, P.; Xu, X. BcCFEM1, a CFEM Domain-Containing Protein with Putative GPI-Anchored Site, Is Involved in Pathogenicity, Conidial Production, and Stress Tolerance in Botrytis cinerea. Front. Microbiol. 2017, 8, 1807. [Google Scholar] [CrossRef]
- De Zwaan, T.M.; Carroll, A.M.; Valent, B.; Sweigard, J.A. Magnaporthe grisea Pth11p Is a Novel Plasma Membrane Protein That Mediates Appressorium Differentiation in Response to Inductive Substrate Cues. Plant Cell 1999, 11, 2013–2030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaknin, Y.; Shadkchan, Y.; Levdansky, E.; Morozov, M.; Romano, J.; Osherov, N. The three Aspergillus fumigatus CFEM-domain GPI-anchored proteins (CfmA-C) affect cell-wall stability but do not play a role in fungal virulence. Fungal Genet. Biol. 2014, 63, 55–64. [Google Scholar] [CrossRef] [PubMed]








| Assigned Name | Target Name | Protein | Length | Total Number of Cysteines | Molecular Weight (kDa) | pI | Predicted Putative Function |
|---|---|---|---|---|---|---|---|
| LsCysRP1 | comp30850_c0_seq1 | MDRLSSMRSVFWILVSFGTNAAVSELINVACPQSAEVDHCMCNCRAIDDAWNVDWHNDLRMFGLKTEIQRPQNEVCCNLVTSCVFIHWDLVSHRKSRNLR | 100 | 7 | 8.9 | 6.04 | No significant similarity found |
| LsCysRP2 | comp28855_c0_seq1 | MGMALFCPVSGLQQQITAATSKFDGKGTSGVGACCTKPCPCQHQMKKLCCISSYGYCNRFRFKELSYHHLVAKANHSWLSCIEVIFAIPYSSADLGWLEMVKP | 103 | 9 | 9.5 | 8.84 | No significant similarity found |
| LsCysRP3 | comp18453_c1_seq1 | MASSGILNLACVVLVSMVLVAPHAEAAMTCGQVTSSVSPCLPYLKGASPLQPGCCNGIRSLNSAAKTTPDRQAACRCLQQAASSISGINLGLASGLPGKCGISI | 104 | 8 | 7.7 | 9.03 | Lipid-transfer protein type 1 |
| LsCysRP4 | comp37785_c0_seq1 | MGFTGMSPMSFLCAWCCGQPTHHLEALVSWTAALLQSLKLKVAEDTDFFTNSFDPVEVWIISLYKYLYHKTLRKTNIGKEAALKRAKPMMIACVVEMLLIDLQS | 104 | 4 | 8.9 | 8.8 | No significant similarity found |
| LsCysRP5 | comp31840_c0_seq3 | MAIYKALLASLLVSLFVLHLVVADGMGMQTTDAATSSPPKSSPPKKIDCGGACSSRCQLSSRPNLCKRACGTCCARCNCVPPGTSGNEDVCPCYASMTTHGGRHKCP | 107 | 12 | 8.6 | 8.67 | Gibberellin-regulated protein 1-like |
| NpCysRP1 | EOD50423.1 | MQFKFALVSLFTALALAAPGIDLEKRCTANGGSCTQLSQCCSGNCEYDSSIGLVCKAASKKEKRCTANGGACTQLAQCCSGNCEYDSSIGLVCKP | 95 | 12 | 7.8 | 7.57 | Hypothetical protein UCRNP2_2803 |
| NpCysRP2 | EOD50922.1 | MRPLYSHLVHTLPPLFSILLDPRPASAQDDHVPRCYYPDGTLASNDYACRLNTTESFCCTTNVTCLDNKICQVLAPTQYEYNRGTCTDKTWTSDECPKFCQAQSPSYGSGVIRPVY | 116 | 9 | 10 | 4.79 | Hypothetical protein UCRNP2_2308 |
| NpCysRP3 | EOD52838.1 | MRFFVAISVAATALFSTGLAASHAKCACQIASDRGTNDAATAGMCSYVGGVMSKSNVAFEKQVATWSLTSILADNTITTGVLYPGKYCQGIGGHELGGDAVYEACRSQKCGDTYCQDSTCV | 121 | 8 | 10.5 | 5.48 | Hypothetical protein UCRNP2_338 |
| NpCysRP4 | EOD51105 | MRVSAFAAILGYSSLALAQTQLLPTCAQKCVGTDFGGCSTVDVKCICANKELLTGLACCVSTGCNAADQETVINFAQNLCKPQGVTDLPTTATCASGASSATSSASSGTASTTASSAASASASATGSAASSASSAASSVASSAASAASSAAASASAATTAGSGAGSCQGNAAGMGFGLVLAGLLGAL | 187 | 11 | 15.5 | 4.53 | Putative CFEM domain protein |
| NpCysRP5 | EOD44996.1 | MKTSFAAVAFSLASVAYSQNISDLPSCSLNCFVTTLGSDGCSSLTDFECHCKVPGLTDEITPCVQKACSAADQQTVAEQVQALCAQYGVSISVPEASSTAAPSSTAAATSAPASSSAAATTAASSAASSAASSASASATSTETASPSATDVVISTPVATPTTPGASTPAGSTPTPSQFQGAAGKTGVVGGVVGFAAAVAAVAAL | 204 | 8 | 17.6 | 4.02 | Putative CFEM domain protein |
| Assigned Name | TargetP-2.0 Server Prediction | Cleavage Site between Pos | Likelihood | Protter | Cleavage Site between Pos | DeepLoc-1.0 Prediction | Likelihood | Soluble/Membrane |
|---|---|---|---|---|---|---|---|---|
| LsCysP1 | Signal peptide | 24–25 | 0.9931 | Signal peptide | 21–22 | Extracellular | 0.8321 | 0.9813/0.0187 |
| LsCysP2 | Other | ⎻⎻⎻⎻ | 0.7782 | Signal peptide | 18–19 | Extracellular | 0.8088 | 0.9946/0.0054 |
| LsCysP3 | Signal peptide | 26–27 | 1 | Signal peptide | 26–27 | Extracellular | 0.9998 | 1/0 |
| LsCysP4 | Signal peptide | 26–27 | 0.7135 | Signal peptide | 26–27 | Extracellular | 0.7316 | 0.9415/0.0585 |
| LsCysP5 | Signal peptide | 23–24 | 1 | Signal peptide | 23–24 | Extracellular | 0.9973 | 1/0 |
| NpCysP1 | Signal peptide | 17–18 | 1 | Signal peptide | 20–21 | Extracellular | 1 | 1/0 |
| NpCysP2 | Signal peptide | 27–28 | 0.9962 | Signal peptide | 27–28 | Extracellular | 0.8773 | 0.9997/0.0003 |
| NpCysP3 | Signal peptide | 20–21 | 1 | Signal peptide | 20–21 | Extracellular | 0.999 | 0.998/0.002 |
| NpCysP4 | Signal peptide | 18–19 | 0.999 | Signal peptide | 18–19 | cell membrane/extracellular/Endoplasmic reticulum | 0.2925/0.2815 /0.2539 | 0.3246/0.6754 |
| NpCysP5 | Signal peptide | 18–19 | 0.9986 | Signal peptide | 18–19 | Extracellular/Endoplasmic reticulum/Cell membrane | 0.3539/0.2639 /.2186 | 0.4355/0.5645 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vázquez-Avendaño, R.; Rodríguez-Haas, J.B.; Velázquez-Delgado, H.; Rosas-Saito, G.H.; Hernández-Domínguez, E.E.; Sánchez-Rangel, D. Insights of the Neofusicoccum parvum–Liquidambar styraciflua Interaction and Identification of New Cysteine-Rich Proteins in Both Species. J. Fungi 2021, 7, 1027. https://doi.org/10.3390/jof7121027
Vázquez-Avendaño R, Rodríguez-Haas JB, Velázquez-Delgado H, Rosas-Saito GH, Hernández-Domínguez EE, Sánchez-Rangel D. Insights of the Neofusicoccum parvum–Liquidambar styraciflua Interaction and Identification of New Cysteine-Rich Proteins in Both Species. Journal of Fungi. 2021; 7(12):1027. https://doi.org/10.3390/jof7121027
Chicago/Turabian StyleVázquez-Avendaño, Rebeca, José Benjamín Rodríguez-Haas, Hugo Velázquez-Delgado, Greta Hanako Rosas-Saito, Eric Edmundo Hernández-Domínguez, and Diana Sánchez-Rangel. 2021. "Insights of the Neofusicoccum parvum–Liquidambar styraciflua Interaction and Identification of New Cysteine-Rich Proteins in Both Species" Journal of Fungi 7, no. 12: 1027. https://doi.org/10.3390/jof7121027
APA StyleVázquez-Avendaño, R., Rodríguez-Haas, J. B., Velázquez-Delgado, H., Rosas-Saito, G. H., Hernández-Domínguez, E. E., & Sánchez-Rangel, D. (2021). Insights of the Neofusicoccum parvum–Liquidambar styraciflua Interaction and Identification of New Cysteine-Rich Proteins in Both Species. Journal of Fungi, 7(12), 1027. https://doi.org/10.3390/jof7121027