Applying EDTA in Chelating Excess Metal Ions to Improve Downstream DNA Recovery from Mine Tailings for Long-Read Amplicon Sequencing of Acidophilic Fungi Communities
Abstract
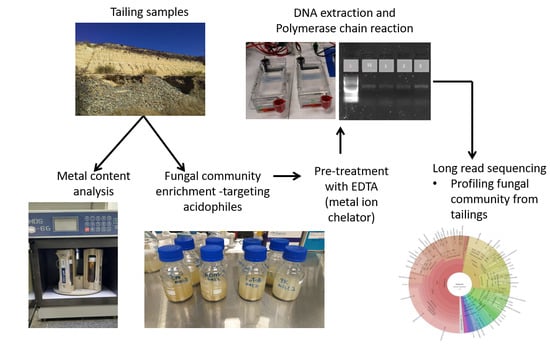
:1. Introduction
2. Materials and Methods
2.1. Site Description and Soil Sampling
2.2. Characterization of Mine Tailing Samples
2.3. Microbial Profiling and Isolation
2.3.1. Enrichment and DNA Extraction
- Solution A contains all the compounds listed in Table 1 (for each media) except iron (II) sulfate or sulfur, which were sterilized by autoclaving at 112 °C for 30 min.
- Solution B contains either iron (II) sulfate (44.2 g/L in liquid media and 6 g/L on solid media, maintained at pH 2 to avoid oxidation) or sulfur (30 g/L in liquid media and 10 g/L) [26,27]. The solutions were sterilized by filtration using 0.2 µm sterile filters and added to solution A aseptically to prepare iron- or sulfur-containing media [28].
2.3.2. Optimization of DNA Extraction
2.3.3. Bioinformatics and Statistics
3. Results
3.1. Tailing Characterization
3.2. DNA Optimization
3.3. Alpha and Beta Diversities
3.4. Taxonomic Diversity of Microbial Community on GYEM and YSM Enrichment
4. Discussion
4.1. DNA Extraction Optimization
4.2. Taxonomic Diversity of Microbial Community in Tailing Samples
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vázquez-Campos, X.; Kinsela, A.S.; Waite, T.D.; Collins, R.N.; Neilan, B.A. Fodinomyces uranophilus gen. nov. sp. nov. and Coniochaeta fodinicola sp. nov., two uranium mine-inhabiting Ascomycota fungi from northern Australia. Mycologia 2014, 106, 1073–1089. [Google Scholar] [CrossRef]
- Liang, J.; Liu, J.; Yang, T.; Wang, P.D.; Zhang, S.C.; Jia, P.; Liao, B.; Shu, W.S.; Li, J.T. Contrasting soil fungal communities at different habitats in a revegetated copper mine wasteland. Soil Ecol. Lett. 2020, 2, 8–19. [Google Scholar] [CrossRef] [Green Version]
- Jia, T.; Wang, R.; Fan, X.; Chai, B. A comparative study of fungal community structure, diversity and richness between the soil and the phyllosphere of native grass species in a copper tailings dam in Shanxi Province, China. Appl. Sci. 2018, 8, 1297. [Google Scholar] [CrossRef] [Green Version]
- Huerta-Rosas, B.; Cano-Rodríguez, I.; Gamiño-Arroyo, Z.; Gómez-Castro, F.I.; Carrillo-Pedroza, F.R.; Romo-Rodríguez, P.; Gutiérrez-Corona, J.F. Aerobic processes for bioleaching manganese and silver using microorganisms indigenous to mine tailings. World J. Microbiol. Biotechnol. 2020, 36, 124. [Google Scholar] [CrossRef]
- Selbmann, L.; Stoppiello, G.A.; Onofri, S.; Stajich, J.E. Culture-Dependent and Amplicon Sequencing Approaches Reveal Diversity and Distribution of Black Fungi in Antarctic Cryptoendolithic Communities. J. Fungi 2021, 7, 213. [Google Scholar] [CrossRef]
- Desai, C.; Madamwar, D. Extraction of inhibitor-free metagenomic DNA from polluted sediments, compatible with molecular diversity analysis using adsorption and ion-exchange treatments. Bioresour. Technol. 2007, 98, 761–768. [Google Scholar] [CrossRef]
- Liu, M.; Ding, X.; Wang, X.; Li, J.; Yang, H.; Yin, Y. Extraction of DNA from complex biological sample matrices using guanidinium ionic liquid modified magnetic nanocomposites. RSC Adv. 2019, 9, 23119–23128. [Google Scholar] [CrossRef] [Green Version]
- Colin, Y.; Goberna, M.; Verdú, M.; Navarro-Cano, J.A. Successional trajectories of soil bacterial communities in mine tailings: The role of plant functional traits. J. Environ. Manag. 2019, 241, 284–292. [Google Scholar] [CrossRef]
- Bernardino, A.F.; Pais, F.S.; Oliveira, L.S.; Gabriel, F.A.; Ferreira, T.O.; Queiroz, H.M.; Mazzuco, A.C.A. Chronic trace metals effects of mine tailings on estuarine assemblages revealed by environmental DNA. PeerJ 2019, 2019, e8042. [Google Scholar] [CrossRef]
- Kuffel, A.; Gray, A.; Daeid, N.N. Impact of metal ions on PCR inhibition and RT-PCR efficiency. Int. J. Legal Med. 2021, 135, 63–72. [Google Scholar] [CrossRef]
- Moreno, L.I.; McCord, B.R. Understanding metal inhibition: The effect of copper (Cu2+) on DNA containing samples. Forensic Chem. 2017, 4, 89–95. [Google Scholar] [CrossRef]
- Guéroult, M.; Picot, D.; Abi-Ghanem, J.; Hartmann, B.; Baaden, M. How cations can assist dnase i in DNA binding and hydrolysis. PLoS Comput. Biol. 2010, 6, e1001000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR inhibitors—Occurrence, properties and removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, K.; Ward, W.S. A novel nuclease activity that is activated by Ca2+ chelated to EGTA. Syst. Biol. Reprod. Med. 2009, 55, 193–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melgar, E.; Goldthwait, D.A. Deoxyribonucleic acid nucleases. II. The effects of metals on the mechanism of action of deoxyribonuclease I. J. Biol. Chem. 1968, 243, 4409–4416. [Google Scholar] [CrossRef]
- El-Ashram, S.; Al Nasr, I.; Suo, X. Nucleic acid protocols: Extraction and optimization. Biotechnol. Rep. 2016, 12, 33–39. [Google Scholar] [CrossRef] [Green Version]
- Heikrujam, J.; Kishor, R.; Behari Mazumder, P. The Chemistry Behind Plant DNA Isolation Protocols. Biochem. Anal. Tools–Methods Bio-Molecules Stud. 2020, 8, 131–141. [Google Scholar] [CrossRef]
- Bonsu, D.O.M.; Higgins, D.; Austin, J.J. Forensic touch DNA recovery from metal surfaces—A review. Sci. Justice 2020, 60, 206–215. [Google Scholar] [CrossRef]
- Feng, D.; van Deventer, J.S.J. Thiosulphate leaching of gold in the presence of ethylenediaminetetraacetic acid (EDTA). Miner. Eng. 2010, 23, 143–150. [Google Scholar] [CrossRef]
- Lopata, A.; Jójárt, B.; Surányi, É.V.; Takács, E.; Bezúr, L.; Leveles, I.; Bendes, Á.; Viskolcz, B.; Vértessy, B.G.; Tóth, J. Beyond chelation: EDTA tightly binds taq DNA polymerase, MutT and dUTPase and directly inhibits dNTPase activity. Biomolecules 2019, 9, 621. [Google Scholar] [CrossRef] [Green Version]
- Portzehl, H.; Caldwell, P.C.; Rüegg, J.C. The dependence of contraction and relaxation of muscle fibres from the crab Maia squinado on the internal concentration of free calcium ions. BBA-Spec. Sect. Biophys. Subj. 1964, 79, 581–591. [Google Scholar] [CrossRef]
- Zaitoun, M.A.; Lin, C.T. Chelating behavior between metal ions and EDTA in sol—gel matrix. J. Phys. Chem. B 1997, 101, 1857–1860. [Google Scholar] [CrossRef]
- Khosravinia, H.; Ramesha, K.P. Influence of EDTA and magnesium on DNA extraction from blood samples and specificity of polymerase chain reaction. Afr. J. Biotechnol. 2007, 6, 184–187. [Google Scholar] [CrossRef]
- Hall, N.E.L.; Axelrod, D.E. Interference of cellular ferric ions with DNA extraction and the application to methods of DNA determination. Anal. Biochem. 1977, 79, 425–430. [Google Scholar] [CrossRef]
- Tang, C.; Zhong, J.; Lv, Y.; Liu, X.; Li, Y.; Zhang, M.; Yan, X.; Sun, W. Response and dynamic change of microbial community during bioremediation of uranium tailings by bacillus sp. Minerals 2021, 11, 967. [Google Scholar] [CrossRef]
- Hassanshahian, M.; Ghoebani, S. Isolation and Characterization of Iron and Sulfur- Oxidizing Bacteria from Maiduk Copper Mine at Shahrbabk Province in Iran. Geomicrobiol. J. 2018, 35, 261–265. [Google Scholar] [CrossRef]
- Martínez, P.; Gálvez, S.; Ohtsuka, N.; Budinich, M.; Cortés, M.P.; Serpell, C.; Nakahigashi, K.; Hirayama, A.; Tomita, M.; Soga, T.; et al. Metabolomic study of Chilean biomining bacteria Acidithiobacillus ferrooxidans strain Wenelen and Acidithiobacillus thiooxidans strain Licanantay. Metabolomics 2012, 9, 247–257. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.; Lin, H.; Xu, X.; Zhou, S. Hydrometallurgy Bioleaching of different copper sul fi des by Acidithiobacillus ferrooxidans and its adsorption on minerals. Hydrometallurgy 2013, 140, 42–47. [Google Scholar] [CrossRef]
- Kishimoto, N.; Tano, T. Acidophilic heterotrophic bacteria isolated from acidic mine drainage, sewage, and soils. J. Gen. Appl. Microbiol. 1987, 33, 11–25. [Google Scholar] [CrossRef] [Green Version]
- Amiri, F.; Mousavi, S.M.; Yaghmaei, S. Enhancement of bioleaching of a spent Ni/Mo hydroprocessing catalyst by Penicillium simplicissimum. Sep. Purif. Technol. 2011, 80, 566–576. [Google Scholar] [CrossRef]
- Rasheed, A.; Cloete, E.; Khasa, D. Isolation and identification of iron ore—Solubilising fungus. S. Afr. J. Sci. 2010, 106, 1–6. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2 ITS Pipeline Workflow (1.8). 2020. Available online: https://benjjneb.github.io/dada2/ITS_workflow.html (accessed on 22 November 2021).
- Callahan, B.J.; Wong, J.; Heiner, C.; Oh, S.; Theriot, C.M.; Gulati, A.S.; McGill, S.K.; Dougherty, M.K. High-throughput amplicon sequencing of the full-length 16S rRNA gene with single-nucleotide resolution. Nucleic Acids Res. 2019, 47, e103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanunchai, B.; Ji, L.; Schroeter, S.A.; Wahdan, S.F.M.; Hossen, S.; Delelegn, Y.; Buscot, F.; Lehnert, A.S.; Alves, E.G.; Hilke, I.; et al. FungalTraits vs. FUNGuild: Comparison of Ecological Functional Assignments of Leaf- and Needle-Associated Fungi across 12 Temperate Tree Species. Microb. Ecol. 2022, 1–18. [Google Scholar] [CrossRef]
- Põlme, S.; Abarenkov, K.; Henrik Nilsson, R.; Lindahl, B.D.; Clemmensen, K.E.; Kauserud, H.; Nguyen, N.; Kjøller, R.; Bates, S.T.; Baldrian, P.; et al. FungalTraits: A user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Divers. 2020, 105, 1–16. [Google Scholar] [CrossRef]
- Kargar, S. Determination of DNA Concentration and Purity by Ultraviolet Spectrophotometry. Academia. Available online: https://www.academia.edu/29266669/Determination_of_DNA_Concentration_and_Purity_by_Ultraviolet_Spectrophotometry (accessed on 7 April 2022).
- Lucena-Aguilar, G.; Sánchez-López, A.M.; Barberán-Aceituno, C.; Carrillo-Ávila, J.A.; López-Guerrero, J.A.; Aguilar-Quesada, R. DNA Source Selection for Downstream Applications Based on DNA Quality Indicators Analysis. Biopreserv. Biobank. 2016, 14, 264–270. [Google Scholar] [CrossRef] [Green Version]
- Xue, P.; Carrillo, Y.; Pino, V.; Minasny, B.; Mcbratney, A.B. Soil Properties Drive Microbial Community Structure in a Large Scale Transect in South Eastern. Sci. Rep. 2018, 8, 11725. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Wang, J.; Lv, Y.; Liu, X.; Zhong, J. Effects of Heavy Metals/Metalloids and Soil Properties on Microbial Communities in Farmland in the Vicinity of a Metals Smelter. Front. Microbiol. 2021, 12, 2347. [Google Scholar] [CrossRef]
- Xia, M.; Bao, P.; Liu, A.; Wang, M.; Shen, L.; Yu, R.; Liu, Y.; Chen, M.; Li, J.; Wu, X.; et al. Bioleaching of low-grade waste printed circuit boards by mixed fungal culture and its community structure analysis. Resour. Conserv. Recycl. 2018, 136, 267–275. [Google Scholar] [CrossRef]
- Begum, S.; Rath, S.K.; Rath, C.C. Applications of Microbial Communities for the Remediation of Industrial and Mining Toxic Metal Waste: A Review. Geomicrobiol. J. 2021, 1–12. [Google Scholar] [CrossRef]
- Selbmann, L.; de Hoog, G.S.; Zucconi, L.; Isola, D.; Ruisi, S.; Gerrits van den Ende, A.H.G.; Ruibal, C.; De Leo, F.; Urzì, C.; Onofri, S. Drought meets acid: Three new genera in a dothidealean clade of extremotolerant fungi. Stud. Mycol. 2008, 61, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Coleine, C.; Stajich, J.E.; Ríos, A.D.L.; Selbmann, L.; Coleine, C.; Stajich, J.E.; Ríos, A.D.L.; Selbmann, L.; Coleine, C.; Stajich, J.E. Beyond the extremes: Rocks as ultimate refuge for fungi in drylands Beyond the extremes: Rocks as ultimate refuge for fungi in drylands. Mycologia 2021, 113, 108–133. [Google Scholar] [CrossRef] [PubMed]
- Ametrano, C.G.; Muggia, L.; Grube, M. Fungi in Extreme Environments: Ecological Role and Biotechnological Significance. Fungi Extrem. Environ. Ecol. Role Biotechnol. Significance 2019, 119–143. [Google Scholar] [CrossRef]
- Lombard, L.; van der Merwe, N.A.; Groenewald, J.Z.; Crous, P.W. Generic concepts in Nectriaceae. Stud. Mycol. 2015, 80, 189–245. [Google Scholar] [CrossRef] [Green Version]
- Patel, J.N.; Parmar, F.A.; Upasani, U.V. Isolation and Characterization of Pathogens Causing Disease in Pomegranate (Punica granatum L.), India. IJIRSET 2021, 10. [Google Scholar] [CrossRef]
- Taha, H.; Shivanand, P.; Khoo, D.H.; Mohammad, Y.H.; Matussin, N.B.A.; Metali, F. Identification of culturable petroleum-degrading bacteria and fungi from petroleum-contaminated sites in Brunei Darussalam. J. Environ. Sci. Health-Part A Toxic/Hazardous Subst. Environ. Eng. 2020, 55, 1542–1547. [Google Scholar] [CrossRef]
- Hujslová, M.; Kubátová, A.; Kostovčík, M.; Blanchette, R.A.; de Beer, Z.W.; Chudíčková, M.; Kolařík, M. Three new genera of fungi from extremely acidic soils. Mycol. Prog. 2014, 13, 819–831. [Google Scholar] [CrossRef] [Green Version]
- Tansengco, M.; Tejano, J.; Coronado, F.; Gacho, C.; Barcelo, J. Heavy metal tolerance and removal capacity of trichoderma species isolated from mine tailings in Itogon, Benguet. Environ. Nat. Resour. J. 2018, 16, 39–57. [Google Scholar] [CrossRef]
- Ou, S.N.; Liang, J.L.; Jiang, X.M.; Liao, B.; Jia, P.; Shu, W.S.; Li, J.T. Physiological, Genomic and Transcriptomic Analyses Reveal the Adaptation Mechanisms of Acidiella bohemica to Extreme Acid Mine Drainage Environments. Front. Microbiol. 2021, 12, 705839. [Google Scholar] [CrossRef]
- An, H.; Liu, Y.; Zhao, X.; Huang, Q.; Yuan, S.; Yang, X.; Dong, J. Characterization of cadmium-resistant endophytic fungi from Salix variegata Franch. in Three Gorges Reservoir Region, China. Microbiol. Res. 2015, 176, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Takano, K.; Itoh, Y.; Ogino, T.; Kurosawa, K.; Sasaki, K. Phylogenetic analysis of manganese-oxidizing fungi isolated from manganese-rich aquatic environments in Hokkaido, Japan. Limnology 2006, 7, 219–223. [Google Scholar] [CrossRef]
- Tanney, J.B.; Seifert, K.A. Mollisiaceae: An overlooked lineage of diverse endophytes. Stud. Mycol. 2020, 95, 293–380. [Google Scholar] [CrossRef] [PubMed]
- Clark, T.N.; Ellsworth, K.T.; Decken, A.; Johnson, J.A.; Gray, C.A. The Application of nmr and lc-hrms Based Prioritization Strategies for the Discovery of Natural Products Produced by Endophytic Fungi from Medicinal Plants. Doctoral Dissertation, University of New Brunswick, Fredericton and Saint John, NB, Canada, 2019. [Google Scholar]
- Zappelini, C.; Karimi, B.; Foulon, J.; Lacercat-Didier, L.; Maillard, F.; Valot, B.; Blaudez, D.; Cazaux, D.; Gilbert, D.; Yergeau, E.; et al. Diversity and complexity of microbial communities from a chlor-alkali tailings dump. Soil Biol. Biochem. 2015, 90, 101–110. [Google Scholar] [CrossRef]
- Bomberg, M.; Miettinen, H.; Musuku, B.; Kinnunen, P. First insights to the microbial communities in the plant process water of the multi-metal Kevitsa mine. Res. Microbiol. 2020, 171, 230–242. [Google Scholar] [CrossRef]
- Miettinen, H.; Bomberg, M.; Le, T.M.K.; Kinnunen, P. Identification and metabolism of naturally prevailing microorganisms in zinc and copper mineral processing. Minerals 2021, 11, 156. [Google Scholar] [CrossRef]
- Sparber, F.; Ruchti, F.; LeibundGut-Landmann, S. Host Immunity to Malassezia in Health and Disease. Front. Cell. Infect. Microbiol. 2020, 10, 198. [Google Scholar] [CrossRef]
- Amend, A. From Dandruff to Deep-Sea Vents: Malassezia-like Fungi Are Ecologically Hyper-diverse. PLoS Pathog. 2014, 10, e1004277. [Google Scholar] [CrossRef] [Green Version]
- Garmendia, G.; Alvarez, A.; Villarreal, R.; Martínez-Silveira, A.; Wisniewski, M.; Vero, S. Fungal diversity in the coastal waters of King George Island (maritime Antarctica). World J. Microbiol. Biotechnol. 2021, 37, 142. [Google Scholar] [CrossRef]
- Samarakoon, B.C.; Phookamsak, R.; Wanasinghe, D.N.; Chomnunti, P.; Hyde, K.D.; McKenzie, E.H.C.; Promputtha, I.; Xu, J.C.; Li, Y.J. Taxonomy and phylogenetic appraisal of Spegazzinia musae sp. nov. And S. deightonii (Didymosphaeriaceae, Pleosporales) on Musaceae from Thailand. MycoKeys 2020, 70, 19–37. [Google Scholar] [CrossRef]
- Mahmoud, R.S.; Narisawa, K. A new fungal endophyte, Scolecobasidium humicola, promotes tomato growth under organic nitrogen. PLoS ONE 2013, 8, e78746. [Google Scholar] [CrossRef] [PubMed]
- Hamayun, M.; Khan, S.A.; Kim, H.Y.; Chaudhary, M.F.; Hwang, Y.H.; Shin, D.H.; Kim, I.K.; Lee, B.H.; Lee, I.J. Gibberellin Production and Plant Growth Enhancement by Newly Isolated Strain of Scolecobasidium tshawytschae. J. Microbiol. Biotechnol. 2009, 19, 560–565. [Google Scholar] [PubMed]





| Name | Composition | ||
|---|---|---|---|
| General | |||
| Basic bioleaching media composition | 3.0 g/L (NH4)2SO4, 0.5 g/L MgSO4·7H2O, 0.5 g/L K2HPO4, 0.1 g/L KCl, 44.2 g/L FeSO4·7H2O or 30 g/L sulfur and 1.5% Agar bacteriological | ||
| Compounds Different in Each Media | pH | References | |
| Glucose yeast extract medium (GYEM) | 5 g/L glucose, 0.05 g/L yeast extract | 3 | [29] |
| Yeast sucrose media (YSM) | 100 g/L sucrose, 1.5 g/L NaNO3, 1.6 g/L yeast extract | 3 | [30] |
| Depths | Mixed Tailings | Tailing A | Tailing B | Tailing C |
|---|---|---|---|---|
| 1: 0–15 cm | 3.27 ± 0.75 | 2.1 ± 0.017 | 3.0 ± 1.06 | 3.945 ± 0.02 |
| 2: 15–30 cm | 3.305 ± 0.46 | 1.8 ± 0.04 | 2.8 ± 1.12 | 3.7 ± 0.04 |
| 3: 30–45 cm | 3.25 ± 0.61 | 1.8 ± 0.005 | 2.8 ± 1.12 | 3.7 ± 0.01 |
| Sample ID | A260/A280 | Fluorometer (ng/mL) | Reads Count | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Glucose Yeast Extract Media | ||||||||||
| S1 | S2 | S1 | S2 | S1 | S2 | |||||
| No EDTA | EDTA | No EDTA | EDTA | No EDTA | EDTA | No EDTA | EDTA | EDTA | EDTA | |
| TM1 | 2.0 | 1.95 | 1.83 | 2.50 | 1.25 | 78.5 | 1.50 | 14.4 | 43,557 | 25,392 |
| TM2 | 1.55 | 2.00 | 2.0 | 2.33 | 1.18 | 1.09 | 2.00 | 2.08 | 45,882 | 43,225 |
| TM3 | 1.40 | 2.00 | 1.50 | 2.66 | low | 0.59 | 2.50 | 14.5 | 10,319 | 76,036 |
| 1TA | 1.60 | 2.10 | 2.10 | 2.40 | 1.31 | 140 | 0.58 | 19.6 | 60,803 | 3868 |
| 2TA | 1.5 | 2.00 | 1.93 | 2.70 | 0.98 | 172 | 1.95 | 7.59 | 19,980 | 6725 |
| 3TA | 2.0 | 2.00 | 1.55 | 2.75 | 1.51 | 6.64 | 0.88 | 7.72 | 60,633 | 6365 |
| 1TB | 1.66 | 1.92 | 2.00 | 2.20 | 0.89 | 225 | 2.80 | 42.1 | 34,189 | 125,058 |
| 2TB | 1.50 | 1.9 | 1.01 | 2.20 | low | 37.8 | 0.94 | 7.61 | 51,553 | 44,624 |
| 3TB | 2.0 | 1.95 | 1.35 | 2.20 | 0.84 | 139 | 1.21 | 13.5 | 43,365 | 22,567 |
| 1TC | 1.71 | 2.14 | 1.88 | 2.25 | 1.29 | 8.88 | low | 2.09 | 37,915 | 45,172 |
| 2TC | 1.50 | 2.14 | 1.6 | 1.92 | 1.07 | 26.0 | 3.08 | 340 | 31,218 | 100,441 |
| 3TC | 1.80 | 2.00 | 2.00 | 2.50 | 5.48 | 58 | 0.86 | 5.54 | 7045 | 75,451 |
| Yeast Sucrose Media | ||||||||||
| S1 | S2 | S1 | S2 | S1 | S2 | |||||
| No EDTA | EDTA | No EDTA | EDTA | No EDTA | EDTA | No EDTA | EDTA | EDTA | EDTA | |
| TM1 | 1.7 | 1.62 | 0.22 | 4.00 | 1.85 | 14.8 | low | 0.75 | 29,426 | 9131 |
| TM2 | 1.54 | 1.50 | 1.40 | 1.50 | 1.47 | 1.54 | low | 0.88 | 27,541 | 681 |
| TM3 | 1.62 | 1.88 | 1.35 | 1.50 | 0.98 | 5.74 | low | 1.32 | 38,058 | failed |
| 1TA | 1.70 | 1.7 | 0.18 | -- | low | 9.31 | low | 0.50 | 22,704 | 568 |
| 2TA | 0.18 | 1.85 | 1.80 | 2.50 | low | 7.79 | low | 2.60 | 23,805 | 33,873 |
| 3TA | 1.27 | 1.87 | 1.44 | 2.25 | low | 183 | low | 3.37 | 24,542 | 20,572 |
| 1TB | 0.90 | 1.78 | 1.35 | 1.36 | low | 64.4 | low | 2.06 | 34,033 | 1193 |
| 2TB | 1.44 | 1.62 | 1.55 | 1.58 | low | 10.1 | low | 3.78 | 6816 | 1433 |
| 3TB | 0.71 | 1.72 | 1.28 | 2.00 | low | 16.5 | low | 2.57 | 11,643 | 1261 |
| 1TC | 1.0 | 1.86 | 0.81 | 2.00 | low | 45.1 | low | 1.19 | 13,849 | 3809 |
| 2TC | 1.40 | 1.93 | 0.23 | -- | low | 9.34 | low | 2.81 | 20,693 | 1685 |
| 3TC | 0.75 | 1.75 | 1.27 | 1.75 | low | 1.30 | low | 1.21 | 27,148 | failed |
| Genera | Primary Lifestyle | Secondary Lifestyle | Comment on Lifestyle | Endophytic Interaction Capability | Plant Pathogenic Capacity | Decay Substrate | Decay Type | Aquatic Habitat | Animal Biotrophic Capacity | Growth Form | Fruitbody Type |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Penicillium | unspecified saprotroph | foliar endophyte | toxin-producing, animal parasite some species, mycoparasite some species | foliar endophyte | 0 | soil | mold | partly aquatic | opportunistic human parasite | filamentous mycelium | 0 |
| Recurvomyces | soil saprotroph | rock-inhabiting | 0 | 0 | 0 | soil | 0 | non-aquatic | 0 | filamentous mycelium | perithecium (hymenium hidden, narrow opening) |
| Talaromyces | unspecified saprotroph | 0 | hypervariable, thermophile, some species animal parasite some species various saprotrophs | 0 | 0 | leaf/fruit/seed, soil, animal material | mold | partly aquatic | opportunistic human parasite | filamentous mycelium | 0 |
| Xenoacremonium | wood saprotroph | 0 | 0 | 0 | 0 | wood | 0 | partly marine (partly non-aquatic) | opportunistic human parasite | filamentous mycelium | 0 |
| Rhodotorula | unspecified saprotroph | foliar endophyte | 0 | foliar endophyte | 0 | 0 | 0 | partly aquatic | opportunistic human parasite | yeast | none |
| Acidothrix | soil saprotroph | 0 | 0 | 0 | 0 | soil | 0 | non-aquatic | 0 | filamentous mycelium | none |
| Malassezia | soil saprotroph | root-associated | 0 | root-associated | root-associated | roots, soil | 0 | partly aquatic | animal-associated/opportunistic human parasite | yeast | none |
| Spegazzinia | wood saprotroph | 0 | 0 | no endophytic capacity | 0 | wood | 0 | non-aquatic | 0 | filamentous mycelium | perithecium (hymenium hidden, narrow opening) |
| Coniochaeta | unspecified saprotroph | foliar endophyte | hypervariable | foliar endophyte | leaf/fruit/seed pathogen | leaf/fruit/seed, soil, dung, animal material | 0 | non-aquatic | animal parasite | filamentous mycelium | cleistothecium (closed, spherical) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nkuna, R.; Ijoma, G.N.; Matambo, T.S. Applying EDTA in Chelating Excess Metal Ions to Improve Downstream DNA Recovery from Mine Tailings for Long-Read Amplicon Sequencing of Acidophilic Fungi Communities. J. Fungi 2022, 8, 419. https://doi.org/10.3390/jof8050419
Nkuna R, Ijoma GN, Matambo TS. Applying EDTA in Chelating Excess Metal Ions to Improve Downstream DNA Recovery from Mine Tailings for Long-Read Amplicon Sequencing of Acidophilic Fungi Communities. Journal of Fungi. 2022; 8(5):419. https://doi.org/10.3390/jof8050419
Chicago/Turabian StyleNkuna, Rosina, Grace N. Ijoma, and Tonderayi S. Matambo. 2022. "Applying EDTA in Chelating Excess Metal Ions to Improve Downstream DNA Recovery from Mine Tailings for Long-Read Amplicon Sequencing of Acidophilic Fungi Communities" Journal of Fungi 8, no. 5: 419. https://doi.org/10.3390/jof8050419
APA StyleNkuna, R., Ijoma, G. N., & Matambo, T. S. (2022). Applying EDTA in Chelating Excess Metal Ions to Improve Downstream DNA Recovery from Mine Tailings for Long-Read Amplicon Sequencing of Acidophilic Fungi Communities. Journal of Fungi, 8(5), 419. https://doi.org/10.3390/jof8050419