Global Analysis of RNA-Dependent RNA Polymerase-Dependent Small RNAs Reveals New Substrates and Functions for These Proteins and SGS3 in Arabidopsis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Library Preparation
2.3. Sequencing Read Processing and Alignment
2.4. Read Count Computation
2.5. Differential Abundance Analysis and RDR Substrate Calling
2.6. Size Classification of smRNAs
2.7. Gene Ontology (GO) Analysis
2.8. Target Prediction and Analysis
2.9. ABA treatment
2.10. NaCl Treatment
2.11. PEG8000 Treatment
3. Results
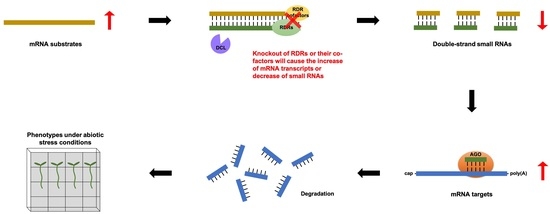
3.1. Analysis of Putative Arabidopsis RDR Substrates
3.2. Small RNA Breakdown Associated with Each Arabidopsis RDR
3.3. Total RNA-seq Data Revealed New Putative Functions for RDRs
3.4. The rdr6 and sgs3 Mutant Plants were Hypersensitive to ABA and Less Sensitive to Salt and PEG8000 Treatment
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mekonnen, M.M.; Hoekstra, A.Y. Four billion people facing severe water scarcity. Sci. Adv. 2016, 2, e1500323. [Google Scholar] [CrossRef] [Green Version]
- Gerland, P.; Raftery, A.E.; Ševčíková, H.; Li, N.; Gu, D.; Spoorenberg, T.; Alkema, L.; Fosdick, B.K.; Chunn, J.; Lalic, N.; et al. World population stabilization unlikely this century. Science 2014, 346, 234–237. [Google Scholar] [CrossRef] [Green Version]
- Finkelstein, R.R.; Gampala, S.; Rock, C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell 2002, 14, S15–S45. [Google Scholar] [CrossRef] [Green Version]
- Seki, M.; Ishida, J.; Narusaka, M.; Fujita, M.; Nanjo, T.; Umezawa, T.; Kamiya, A.; Nakajima, M.; Enju, A.; Sakurai, T.; et al. Monitoring the expression pattern of around 7000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct. Integr. Genom. 2002, 2, 282–291. [Google Scholar] [CrossRef]
- Zhang, J.F.; Yuan, L.J.; Shao, Y.; Du, W.; Yan, D.W.; Lu, Y.T. The disturbance of small RNA pathways enhanced abscisic acid re-sponse and multiple stress responses in Arabidopsis. Plant Cell Environ. 2008, 31, 562–574. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Johansen, L.K.; Gustafson, A.M.; Kasschau, K.D.; Lellis, A.D.; Zilberman, D.; Jacobsen, S.E.; Carrington, J.C. Genetic and Functional Diversification of Small RNA Pathways in Plants. PLoS Biol. 2004, 2, e104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamthan, A.; Chaudhuri, A.; Kamthan, M.; Datt, A. Small RNAs in plants: Recent development and application for crop im-provement. Front. Plant Sci. 2015, 6, 208. [Google Scholar] [CrossRef] [Green Version]
- Baulcombe, D.C. RNA silencing in plants. Nat. Cell Biol. 2004, 431, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matzke, M.A.; Kanno, T.; Matzke, A.J. RNA-Directed DNA Methylation: The Evolution of a Complex Epigenetic Pathway in Flowering Plants. Annu. Rev. Plant Biol. 2015, 66, 243–267. [Google Scholar] [CrossRef] [PubMed]
- Peragine, A.; Yoshikawa, M.; Wu, G.; Albrecht, H.L.; Poethig, R.S. SGS3 and SGS2/SDE1/RDR6 are required for juvenile de-velopment and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004, 19, 2368–2379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willmann, M.R.; Endres, M.W.; Cook, R.T.; Gregory, B.D. The Functions of RNA-Dependent RNA Polymerases in Arabidopsis. Arab. Book 2011, 9, e0146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wassenegger, M.; Krczal, G. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 2006, 11, 142–151. [Google Scholar] [CrossRef]
- Donaire, L.; Barajas, D.; Martínez-García, B.; Martínez-Priego, L.; Pagán, I.; Llave, C. Structural and Genetic Requirements for the Biogenesis of Tobacco Rattle Virus-Derived Small Interfering RNAs. J. Virol. 2008, 82, 5167–5177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.B.; Wu, Q.; Ito, T.; Cillo, F.; Li, W.X.; Chen, X.; Yu, J.L.; Ding, S.W. RNAi-mediated viral immunity requires amplifica-tion of virus-derived siRNAs in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 484–489. [Google Scholar] [CrossRef] [Green Version]
- Olmedo-Monfil, V.; Durán-Figueroa, N.; Arteaga-Vázquez, M.; Demesa-Arévalo, E.; Autran, D.; Grimanelli, D.; Slotkin, R.K.; Martienssen, R.A.; Vielle-Calzada, J.-P. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nat. Cell Biol. 2010, 464, 628–632. [Google Scholar] [CrossRef]
- Finke, A.; Kuhlmann, M.; Mette, M.F. IDN2 has a role downstream of siRNA formation in RNA-directed DNA methylation. Epigenetics 2012, 7, 950–960. [Google Scholar] [CrossRef] [Green Version]
- Verlaan, M.G.; Hutton, S.F.; Ibrahem, R.M.; Kormelink, R.; Visser, R.G.F.; Scott, J.W.; Edwards, J.D.; Bai, Y. The Tomato Yellow Leaf Curl Virus Resistance Genes Ty-1 and Ty-3 Are Allelic and Code for DFDGD-Class RNA–Dependent RNA Polymerases. PLoS Genet. 2013, 9, e1003399. [Google Scholar] [CrossRef] [Green Version]
- Borsani, O.; Zhu, J.; Verslues, P.E.; Sunkar, R.; Zhu, J.K. Endogenous siRNAs derived from a pair of natural cis-antisense tran-scripts regulate salt tolerance in Arabidopsis. Cell 2005, 123, 1279–1291. [Google Scholar] [CrossRef] [Green Version]
- Axtell, M.J.; Jan, C.; Rajagopalan, R.; Bartel, D.P. A Two-Hit Trigger for siRNA Biogenesis in Plants. Cell 2006, 127, 565–577. [Google Scholar] [CrossRef] [Green Version]
- Montgomery, T.A.; Howell, M.D.; Cuperus, J.T.; Li, D.; Hansen, J.E.; Alexander, A.L.; Chapman, E.J.; Fahlgren, N.; Allen, E.; Carrington, J.C. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA for-mation. Cell 2008, 133, 128–141. [Google Scholar] [CrossRef] [Green Version]
- Fahlgren, N.; Montgomery, T.A.; Howell, M.D.; Allen, E.; Dvorak, S.K.; Alexander, A.L.; Carrington, J.C. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 2006, 16, 939–944. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Trudeau, V.L. The XS domain of a plant specific SGS3 protein adopts a unique RNA recognition motif (RRM) fold. Cell Cycle 2008, 7, 2268–2270. [Google Scholar] [CrossRef]
- Bateman, A. The SGS3 protein involved in PTGS finds a family. BMC Bioinform. 2002, 3, 21. [Google Scholar] [CrossRef] [Green Version]
- De Felippes, F.F.; Marchais, A.; Sarazin, A.; Oberlin, S.; Voinnet, O. A single miR390 targeting event is sufficient for triggering TAS3-tasiRNA biogenesis in Arabidopsis. Nucleic Acids Res. 2017, 45, 5539–5554. [Google Scholar] [CrossRef]
- Yoshikawa, M.; Iki, T.; Tsutsui, Y.; Miyashita, K.; Poethig, R.S.; Habu, Y.; Ishikawa, M. 3′ fragment of miR173-programmed RISC-cleaved RNA is protected from degradation in a complex with RISC and SGS3. Proc. Natl. Acad. Sci. USA 2013, 110, 4117–4122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jouannet, V.; Moreno, A.B.; Elmayan, T.; Vaucheret, H.; Crespi, M.D.; Maizel, A. Cytoplasmic Arabidopsis AGO7 accumulates in membrane-associated siRNA bodies and is required for ta-siRNA biogenesis. EMBO J. 2012, 31, 1704–1713. [Google Scholar] [CrossRef]
- Lam, P.; Zhao, L.; McFarlane, H.E.; Aiga, M.; Lam, V.; Hooker, T.S.; Kunst, L. RDR1 and SGS3, Components of RNA-Mediated Gene Silencing, Are Required for the Regulation of Cuticular Wax Biosynthesis in Developing Inflorescence Stems of Arabidopsis. Plant Physiol. 2012, 159, 1385–1395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Q.; Ryvkin, P.; Li, F.; Dragomir, I.; Valladares, O.; Yang, J.; Cao, K.; Wang, L.; Gregory, B. Genome-wide double-stranded RNA sequencing reveals the functional significance of base-paired RNAs in Arabidopsis. PLoS Genet. 2010, 6, e1001141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.; Willmann, M.R.; Vandivier, L.E.; Trefely, S.; Kramer, M.C.; Shapiro, J.; Guo, R.; Lyons, E.; Snyder, N.W.; Gregory, B.D. Messenger RNA 5′ NAD+ Capping is a Dynamic Regulatory Epitranscriptome Mark that is Required for Proper Response to Abscisic Acid in Arabidopsis. SSRN Electron. J. 2020, 56, 125–140. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Du, Z.; Zhou, X.; Ling, Y.; Zhang, Z.; Su, Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010, 38, W64–W70. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Zhao, P. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011, 39, W155–W159. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Verslues, P.E.; Zheng, X.; Lee, B.-H.; Zhan, X.; Manabe, Y.; Sokolchik, I.; Zhu, Y.; Dong, C.-H.; Hasegawa, P.M.; et al. HOS10 encodes an R2R3-type MYB transcription factor essential for cold acclimation in plants. Proc. Natl. Acad. Sci. USA 2005, 102, 9966–9971. [Google Scholar] [CrossRef] [Green Version]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Rajjou, L.; Belghazi, M.; Huguet, R.; Robin, C.; Moreau, A.; Job, C.; Job, D. Proteomic Investigation of the Effect of Salicylic Acid on Arabidopsis Seed Germination and Establishment of Early Defense Mechanisms. Plant Physiol. 2006, 141, 910–923. [Google Scholar] [CrossRef] [Green Version]
- García-Cruz, K.V.; García-Ponce, B.; Garay-Arroyo, A.; Sanchez, M.; Ugartechea-Chirino, Y.; Desvoyes, B.; Pacheco-Escobedo, M.A.; Tapia-López, R.; Ransom-Rodríguez, I.; Gutierrez, C.; et al. The MADS-box XAANTAL1 increases proliferation at the Arabidopsis root stem-cell niche and participates in transition to differentiation by regulating cell-cycle components. Ann. Bot. 2016, 118, 787–796. [Google Scholar] [CrossRef] [Green Version]
- Hugouvieux, V.; Kwak, J.M.; Schroeder, J.I. An mRNA cap binding protein, ABH1, modulates early abscisic acid signal trans-duction in Arabidopsis. Cell 2001, 106, 477–487. [Google Scholar] [CrossRef] [Green Version]
- Stein, M.; Dittgen, J.; Sánchez-Rodríguez, C.; Hou, B.-H.; Molina, A.; Schulze-Lefert, P.; Lipka, V.; Somerville, S. Arabidopsis PEN3/PDR8, an ATP Binding Cassette Transporter, Contributes to Nonhost Resistance to Inappropriate Pathogens That Enter by Direct Penetration. Plant Cell 2006, 18, 731–746. [Google Scholar] [CrossRef] [Green Version]
- Si-Ammour, A.; Windels, D.; Arn-Bouldoires, E.; Kutter, C.; Ailhas, J.; Meins, F., Jr.; Vazquez, F. miR393 and secondary siRNAs regulate expression of the TIR1/AFB2 auxin receptor clade and auxin-related development of Arabidopsis leaves. Plant Physiol. 2011, 157, 683–691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howell, M.D.; Fahlgren, N.; Chapman, E.J.; Cumbie, J.S.; Sullivan, C.M.; Givan, S.A.; Kasschau, K.D.; Carrington, J.C. Genome-Wide Analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 Pathway in Arabidopsis Reveals Dependency on miRNA- and tasiRNA-Directed Targeting. Plant Cell 2007, 19, 926–942. [Google Scholar] [CrossRef] [Green Version]
- Engstrom, E.M.; Andersen, C.M.; Gumulak-Smith, J.; Hu, J.; Orlova, E.; Sozzani, R.; Bowman, J.L. Arabidopsis Homologs of the Petunia HAIRY MERISTEM Gene Are Required for Maintenance of Shoot and Root Indeterminacy. Plant Physiol. 2011, 155, 735–750. [Google Scholar] [CrossRef] [Green Version]
- Palatnik, J.F.; Allen, E.; Wu, X.; Schommer, C.; Schwab, R.; Carrington, J.C.; Weigel, D. Control of leaf morphogenesis by mi-croRNAs. Nature 2003, 425, 257–263. [Google Scholar] [CrossRef] [Green Version]
- Schommer, C.; Palatnik, J.F.; Aggarwal, P.; Chételat, A.; Cubas, P.; Farmer, E.E.; Nath, U.; Weigel, D. Control of Jasmonate Biosynthesis and Senescence by miR319 Targets. PLoS Biol. 2008, 6, e230. [Google Scholar] [CrossRef] [Green Version]
- Tang, G.; Reinhart, B.J.; Bartel, D.P.; Zamore, P.D. A biochemical framework for RNA silencing in plants. Genes Dev. 2003, 17, 49–63. [Google Scholar] [CrossRef] [Green Version]
- Chi, W.T.; Fung, R.; Liu, H.C.; Hsu, C.C.; Charng, Y.Y. Temperature-induced lipocalin is required for basal and acquired ther-motolerance in Arabidopsis. Plant Cell Environ. 2009, 32, 917–927. [Google Scholar] [CrossRef]
- Tiwari, L.D.; Khungar, L.; Grover, A. AtHsc70-1 negatively regulates the basal heat tolerance in Arabidopsis thaliana through affecting the activity of HsfAs and Hsp101. Plant J. 2020, 103, 2069–2083. [Google Scholar] [CrossRef] [PubMed]
- Dufresne, P.J.; Thivierge, K.; Cotton, S.; Beauchemin, C.; Ide, C.; Ubalijoro, E.; Laliberté, J.-F.; Fortin, M.G. Heat shock 70 protein interaction with Turnip mosaic virus RNA-dependent RNA polymerase within virus-induced membrane vesicles. Virology 2008, 374, 217–227. [Google Scholar] [CrossRef] [Green Version]
- Xiong, L.; Schumaker, K.S.; Zhu, J.-K. Cell Signaling during Cold, Drought, and Salt Stress. Plant Cell 2002, 14, S165–S183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michel, B.E.; Porter, J.R.; Sheridan, R.P. Evaluation of the Water Potentials of Solutions of Polyethylene Glycol 8000 Both in the Absence and Presence of Other Solutes. Plant Physiol. 1983, 72, 66–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruggiero, B.; Koiwa, H.; Manabe, Y.; Quist, T.M.; Inan, G.; Saccardo, F.; Joly, R.J.; Hasegawa, P.M.; Bressan, R.A.; Maggio, A. Uncoupling the Effects of Abscisic Acid on Plant Growth and Water Relations. Analysis of sto1/nced3, an Abscisic Acid-Deficient but Salt Stress-Tolerant Mutant in Arabidopsis. Plant Physiol. 2004, 136, 3134–3147. [Google Scholar] [CrossRef] [Green Version]
- Iuchi, S.; Kobayashi, M.; Taji, T.; Naramoto, M.; Seki, M.; Kato, T.; Tabata, S.; Kakubari, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001, 27, 325–333. [Google Scholar] [CrossRef] [Green Version]
- Alzwiy, I.A.; Morris, P.C. A mutation in the Arabidopsis MAP kinase kinase 9 gene results in enhanced seedling stress tolerance. Plant Sci. 2007, 173, 302–308. [Google Scholar] [CrossRef]
- Yoo, S.-D.; Cho, Y.-H.; Tena, G.; Xiong, Y.; Sheen, J. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nat. Cell Biol. 2008, 451, 789–795. [Google Scholar] [CrossRef]
- Polydore, S.; Axtell, M.J. Analysis of RDR1/RDR2/RDR6-independent small RNAs in Arabidopsis thaliana improves MIRNA annotations and reveals unexplained types of short interfering RNA loci. Plant J. 2018, 94, 1051–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsui, A.; Iida, K.; Tanaka, M.; Yamaguchi, K.; Mizuhashi, K.; Kim, J.-M.; Takahashi, S.; Kobayashi, N.; Shigenobu, S.; Shinozaki, K.; et al. Novel Stress-Inducible Antisense RNAs of Protein-Coding Loci Are Synthesized by RNA-Dependent RNA Polymerase. Plant Physiol. 2017, 175, 457–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Genotype | Sense | Antisense | Two Strand | Total |
|---|---|---|---|---|
| rdr1 | 48 | 67 | 21 | 94 |
| rdr2 | 12,488 | 2302 | 2261 | 12,529 |
| rdr3 | 1 | 0 | 0 | 1 |
| rdr4 | 37 | 50 | 24 | 63 |
| rdr5 | 46 | 59 | 26 | 79 |
| rdr6 | 74 | 104 | 54 | 124 |
| sgs3 | 107 | 177 | 65 | 219 |
| rdm12 | 136 | 183 | 72 | 247 |
| rdr1 | rdr2 | rdr3 | rdr4 | rdr5 | rdr6 | sgs3 | |
|---|---|---|---|---|---|---|---|
| rdr2 | 10 | ||||||
| rdr3 | 1 | 1 | |||||
| rdr4 | 7 | 10 | 1 | ||||
| rdr5 | 5 | 13 | 1 | 8 | |||
| rdr6 | 3 | 9 | 0 | 3 | 4 | ||
| sgs3 | 3 | 69 | 0 | 6 | 8 | 27 | |
| rdm12 | 7 | 58 | 1 | 9 | 11 | 5 | 14 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hua, X.; Berkowitz, N.D.; Willmann, M.R.; Yu, X.; Lyons, E.; Gregory, B.D. Global Analysis of RNA-Dependent RNA Polymerase-Dependent Small RNAs Reveals New Substrates and Functions for These Proteins and SGS3 in Arabidopsis. Non-Coding RNA 2021, 7, 28. https://doi.org/10.3390/ncrna7020028
Hua X, Berkowitz ND, Willmann MR, Yu X, Lyons E, Gregory BD. Global Analysis of RNA-Dependent RNA Polymerase-Dependent Small RNAs Reveals New Substrates and Functions for These Proteins and SGS3 in Arabidopsis. Non-Coding RNA. 2021; 7(2):28. https://doi.org/10.3390/ncrna7020028
Chicago/Turabian StyleHua, Xia, Nathan D. Berkowitz, Matthew R. Willmann, Xiang Yu, Eric Lyons, and Brian D. Gregory. 2021. "Global Analysis of RNA-Dependent RNA Polymerase-Dependent Small RNAs Reveals New Substrates and Functions for These Proteins and SGS3 in Arabidopsis" Non-Coding RNA 7, no. 2: 28. https://doi.org/10.3390/ncrna7020028
APA StyleHua, X., Berkowitz, N. D., Willmann, M. R., Yu, X., Lyons, E., & Gregory, B. D. (2021). Global Analysis of RNA-Dependent RNA Polymerase-Dependent Small RNAs Reveals New Substrates and Functions for These Proteins and SGS3 in Arabidopsis. Non-Coding RNA, 7(2), 28. https://doi.org/10.3390/ncrna7020028