The carP lncRNA Is a carS-Related Regulatory Element with Broad Effects on the Fusarium fujikuroi Transcriptome
Abstract
:1. Introduction
2. Results
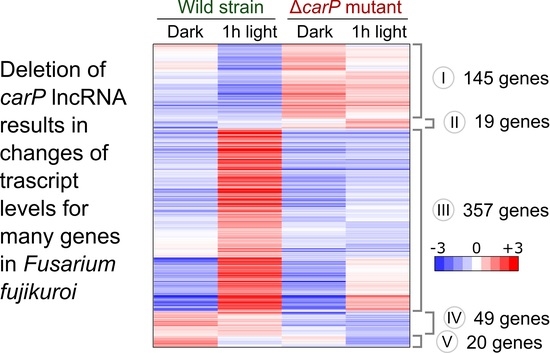
2.1. RNA-seq Analysis of the Effect of ΔcarP Mutation
2.2. Genes Affected by Light in the Wild-Type Strain and in the ΔcarP Mutant
2.3. Genes Differentially Expressed in the ΔcarP Mutant Compared to the Wild-Type Strain
2.4. Reintegration of carP in a ΔcarP Mutant
2.5. Phenotypic Characterization of Transformants Expressing carP in a ΔcarP Mutant
2.6. Relation of the Effects of carP and carS Mutations
3. Discussion
4. Materials and Methods
4.1. Strains and Culture Conditions
4.2. PCR Assays
4.3. Plasmid Constructions and Transformation
4.4. Reintegration of Wild-Type carP in SF268
4.5. Southern Blot
4.6. Expression Studies
4.7. RNA-seq Methodology
4.8. Bioinformatic Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ma, L.J.; Geiser, D.M.; Proctor, R.H.; Rooney, A.P.; O’Donnell, K.; Trail, F.; Gardiner, D.M.; Manners, J.M.; Kazan, K. Fusarium pathogenomics. Annu. Rev. Microbiol. 2013, 67, 399–416. [Google Scholar] [CrossRef] [Green Version]
- Geiser, D.M.; Aoki, T.; Bacon, C.W.; Baker, S.E.; Bhattacharyya, M.K.; Brandt, M.E.; Brown, D.W.; Burgess, L.W.; Chulze, S.; Coleman, J.J.; et al. One fungus, one name: Defining the genus Fusarium in a scientifically robust way that preserves longstanding use. Phytopathology 2013, 103, 400–408. [Google Scholar] [CrossRef] [Green Version]
- Desjardins, A.E.; Proctor, R.H. Molecular biology of Fusarium mycotoxins. Int. J. Food. Microbiol. 2007, 119, 47–50. [Google Scholar] [CrossRef]
- Studt, L.; Tudzynski, B. Gibberellins and the red pigments bikaverin and fusarubin. In Biosynthesis and Molecular Genetics of Fungal Secondary Metabolites; Martín, J.-F., García-Estrada, C., Zeilinger, S., Eds.; Springer: New York, NY, USA, 2014; pp. 209–238. ISBN 978-1-4939-1190-5. [Google Scholar]
- Janevska, S.; Tudzynski, B. Secondary metabolism in Fusarium fujikuroi: Strategies to unravel the function of biosynthetic pathways. Appl. Microbiol. Biotechnol. 2018, 102, 615–630. [Google Scholar] [CrossRef] [PubMed]
- Avalos, J.; Pardo-Medina, J.; Parra-Rivero, O.; Ruger-Herreros, M.; Rodríguez-Ortiz, R.; Hornero-Méndez, D.; Limón, M.C. Carotenoid biosynthesis in Fusarium. J. Fungi 2017, 3, 39. [Google Scholar] [CrossRef] [Green Version]
- Castrillo, M.; Avalos, J. The flavoproteins CryD and VvdA cooperate with the White Collar protein WcoA in the control of photocarotenogenesis in Fusarium fujikuroi. PLoS ONE 2015, 10, e0119785. [Google Scholar] [CrossRef] [Green Version]
- Avalos, J.; Estrada, A.F. Regulation by light in Fusarium. Fungal Genet. Biol. 2010, 47, 930–938. [Google Scholar] [CrossRef] [PubMed]
- García-Martínez, J.; Brunk, M.; Avalos, J.; Terpitz, U. The CarO rhodopsin of the fungus Fusarium fujikuroi is a light-driven proton pump that retards spore germination. Sci. Rep. 2015, 5, 7798. [Google Scholar] [CrossRef] [Green Version]
- Adam, A.; Deimel, S.; Pardo-Medina, J.; García-Martínez, J.; Konte, T.; Limón, M.C.; Avalos, J.; Terpitz, U. Protein activity of the Fusarium fujikuroi rhodopsins CarO and OpsA and their relation to fungus–plant interaction. Int. J. Mol. Sci. 2018, 19, 215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avalos, J.; Cerdá-Olmedo, E. Carotenoid mutants of Gibberella fujikuroi. Curr. Genet. 1987, 11, 505–511. [Google Scholar] [CrossRef]
- Rodríguez-Ortiz, R.; Limón, M.C.; Avalos, J. Functional analysis of the carS gene of Fusarium fujikuroi. Mol. Genet. Genom. 2013, 288, 157–173. [Google Scholar] [CrossRef] [PubMed]
- Ruger-Herreros, M.; Parra-Rivero, O.; Pardo-Medina, J.; Romero-Campero, F.J.; Limón, M.C.; Avalos, J. Comparative transcriptomic analysis unveils interactions between the regulatory CarS protein and light response in Fusarium. BMC Genom. 2019, 20, 67. [Google Scholar] [CrossRef]
- Silva, F.; Navarro, E.; Peñaranda, A.; Murcia-Flores, L.; Torres-Martínez, S.; Garre, V. A RING-finger protein regulates carotenogenesis via proteolysis-independent ubiquitylation of a White Collar-1-like activator. Mol. Microbiol. 2008, 70, 1026–1036. [Google Scholar] [CrossRef]
- Parra-Rivero, O.; Pardo-Medina, J.; Gutiérrez, G.; Limón, M.C.; Avalos, J. A novel lncRNA as a positive regulator of carotenoid biosynthesis in Fusarium. Sci. Rep. 2020, 10, 678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Díaz-Sánchez, V.; Limón, M.C.; Schaub, P.; Al-Babili, S.; Avalos, J. A RALDH-like enzyme involved in Fusarium verticillioides development. Fungal Genet. Biol. 2016, 86, 20–32. [Google Scholar] [CrossRef] [Green Version]
- Pardo-Medina, J.; Gutiérrez, G.; Limón, M.C.; Avalos, J. Impact of the White Collar photoreceptor WcoA on the Fusarium fujikuroi transcriptome. Front. Microbiol. 2021, 11, 619474. [Google Scholar] [CrossRef] [PubMed]
- Cha, J.; Zhou, M.; Liu, Y. Mechanism of the Neurospora circadian clock, a FREQUENCY-centric view. Biochemistry 2015, 54, 150–156. [Google Scholar] [CrossRef] [Green Version]
- Marente, J.; Avalos, J.; Limón, M.C. Controlled transcription of regulator gene carS by Tet-on or by a strong promoter confirms its role as a repressor of carotenoid biosynthesis in Fusarium fujikuroi. Microorganisms 2020, 9, 71. [Google Scholar] [CrossRef]
- Marente, J.; Ortega, P.; Pardo-Medina, J.; Avalos, J.; Limón, M.C. Modulation of activity of a carotenoid pathway through the use of the TET-on regulatory system: Application in the fungus Fusarium fujikuroi. In Plant and Food Carotenoids: Methods and Protocols; Rodríguez-Concepción, M., Welsch, R., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2020; pp. 343–360. ISBN 978-1-4939-9952-1. [Google Scholar]
- Niederer, R.O.; Hass, E.P.; Zappulla, D.C. Long noncoding RNAs in the yeast S. cerevisiae. Adv. Exp. Med. Biol. 2017, 1008, 119–132. [Google Scholar] [CrossRef]
- Martens, J.A.; Laprade, L.; Winston, F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 Gene. Nature 2004, 429, 571–574. [Google Scholar] [CrossRef]
- Houseley, J.; Rubbi, L.; Grunstein, M.; Tollervey, D.; Vogelauer, M. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol. Cell 2008, 32, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Till, P.; Pucher, M.E.; Mach, R.L.; Mach-Aigner, A.R. A long noncoding RNA promotes cellulase expression in Trichoderma reesei. Biotechnol. Biofuels 2018, 11, 78. [Google Scholar] [CrossRef]
- Till, P.; Derntl, C.; Kiesenhofer, D.P.; Mach, R.L.; Yaver, D.; Mach-Aigner, A.R. Regulation of gene expression by the action of a fungal lncRNA on a transactivator. RNA Biol. 2020, 17, 47–61. [Google Scholar] [CrossRef] [PubMed]
- McCluskey, K.; Wiest, A.; Plamann, M. The Fungal Genetics Stock Center: A repository for 50 years of fungal genetics research. J. Biosci. 2010, 35, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Chen, S.; Huang, T.; Zhou, Y.; Han, Y.; Xu, M.; Gu, J. AfterQC: Automatic filtering, trimming, error removing and quality control for Fastq data. BMC Bioinform. 2017, 18, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- de Hoon, M.J.L.; Imoto, S.; Nolan, J.; Miyano, S. Open source clustering software. Bioinformatics 2004, 20, 1453–1454. [Google Scholar] [CrossRef] [Green Version]
- Saldanha, A.J. Java treeview-extensible visualization of microarray data. Bioinformatics 2004, 20, 3246–3248. [Google Scholar] [CrossRef] [Green Version]









| Upregulated 2 | % 3 | Downregulated 2 | % 3 | |
|---|---|---|---|---|
| Light 1 vs. dark in wild-type strain | 970 | 6.4 | 860 | 5.7 |
| Light 1 vs. dark in ΔcarP mutant | 270 | 1.8 | 192 | 1.3 |
| ΔcarP mutant vs. wild-type strain in the dark | 71 | 0.4 | 40 | 0.2 |
| ΔcarP mutant vs. wild-type strain after light 1 | 148 | 1.0 | 405 | 2.7 |
| Primer Set | 5′-3′ Sequence | Experimental Use | |
|---|---|---|---|
| PS1 | Forward | GTAACGCCAGGGTTTTCCCAGTCACACGGCTTGCCAACATGGTGGAGCACGACACTC | neoR segment pRS246neocarP |
| Reverse | TGGATGACGCTTACTATAGTCTTGTCCCAACAAAAGCTGGAGCTCCACCGCGGTGGC | ||
| PS2 | Forward | GCCACCGCGGTGGAGCTCCAGCTTTTGTTGGGACAAGACTATAGTAAGCGTCATCCA | carP segment pRS246neocarP |
| Reverse | GCGGATAACAATTTCACACAGGAAACAGCCAATCCGGGGACAATTCTAGAGGCACGCG | ||
| PS3 | Forward | CGTCGATGCGCCAGTTGATT | carP PCR |
| Reverse | AGCAAGCGCCTAGTGGCC | ||
| PS4 | Forward | CGTCGATGCGCCAGTTGATT | carP-carS PCR |
| Reverse | GTGTAGAGATTGGTGGGGGTT | ||
| PS5 | Forward | CCATTTCTGTTCCCTTCCCTG | carP probe for Southern |
| Reverse | CCGTCATACACCAGAGAGAC | ||
| PS6 | Forward | TCGGTGTCGAGTACCGTCTCT | carB RT-qPCR |
| Reverse | TGCCTTGCCGGTTGCTT | ||
| PS7 | Forward | GATACCCGGCGGAAAGGTTA | carS RT-qPCR |
| Reverse | CTGACAGTCCATTTCAGCGC | ||
| PS8 | Forward | CCATTGAGCTGGGATGTGTTTT | carP RT-qPCR |
| Reverse | TGCGCTGTGCTGTAAACCA | ||
| PS9 | Forward | CCGGTGCTGGAAACAACTG | Reference RT-qPCR (β1-tubulin) |
| Reverse | CGAGGACCTGGTCGACAAGT | ||
| PS10 | Forward | GTGACCTCAAGGGCGTTCTG | Reference RT-qPCR (gpdA) |
| Reverse | CGAAGATGGAGTTTGTGTT | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pardo-Medina, J.; Gutiérrez, G.; Limón, M.C.; Avalos, J. The carP lncRNA Is a carS-Related Regulatory Element with Broad Effects on the Fusarium fujikuroi Transcriptome. Non-Coding RNA 2021, 7, 46. https://doi.org/10.3390/ncrna7030046
Pardo-Medina J, Gutiérrez G, Limón MC, Avalos J. The carP lncRNA Is a carS-Related Regulatory Element with Broad Effects on the Fusarium fujikuroi Transcriptome. Non-Coding RNA. 2021; 7(3):46. https://doi.org/10.3390/ncrna7030046
Chicago/Turabian StylePardo-Medina, Javier, Gabriel Gutiérrez, M. Carmen Limón, and Javier Avalos. 2021. "The carP lncRNA Is a carS-Related Regulatory Element with Broad Effects on the Fusarium fujikuroi Transcriptome" Non-Coding RNA 7, no. 3: 46. https://doi.org/10.3390/ncrna7030046
APA StylePardo-Medina, J., Gutiérrez, G., Limón, M. C., & Avalos, J. (2021). The carP lncRNA Is a carS-Related Regulatory Element with Broad Effects on the Fusarium fujikuroi Transcriptome. Non-Coding RNA, 7(3), 46. https://doi.org/10.3390/ncrna7030046