New Insights into the Roles of lncRNAs as Modulators of Cytoskeleton Architecture and Their Implications in Cellular Homeostasis and in Tumorigenesis
Abstract
:1. Introduction
2. lncRNAs as Cytoskeletal Modulators of Cellular Homeostasis
3. lncRNAs Modulators of Actin Filaments and Accessory Proteins in Cancer Pathogenesis
4. lncRNAs as Modulators of Rho/ROCK Signaling in Tumorigenesis
5. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Elkon, R.; Agami, R. Characterization of noncoding regulatory DNA in the human genome. Nat. Biotechnol. 2017, 35, 732–746. [Google Scholar] [CrossRef] [PubMed]
- García-Padilla, C.; Dueñas, Á.; García-López, V.; Aránega, A.; Franco, D.; Garcia-Martínez, V.; López-Sánchez, C. Molecular Mechanisms of lncRNAs in the Dependent Regulation of Cancer and Their Potential Therapeutic Use. Int. J. Mol. Sci. 2022, 23, 764. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Padilla, C.; Dueñas, A.; Franco, D.; Garcia-Lopez, V.; Aranega, A.; Garcia-Martinez, V.; Lopez-Sanchez, C. Dynamic MicroRNA Expression Profiles during Embryonic Development Provide Novel Insights Into Cardiac Sinus Venosus/Inflow Tract Differentiation. Front. Cell Dev. Biol. 2022, 9, 767954. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 Catalog of Human Long Noncoding RNAs: Analysis of Their Gene Structure, Evolution, and Expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [Green Version]
- De Hoon, M.; Shin, J.W.; Carninci, P. Paradigm shifts in genomics through the FANTOM projects. Mamm. Genome 2015, 26, 391–402. [Google Scholar] [CrossRef] [Green Version]
- Ramilowski, J.A.; Yip, C.W.; Agrawal, S.; Chang, J.-C.; Ciani, Y.; Kulakovskiy, I.V.; Mendez, M.; Ooi, J.L.C.; Ouyang, J.F.; Parkinson, N.; et al. Functional annotation of human long noncoding RNAs via molecular phenotyping. Genome Res. 2020, 30, 1060–1072. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Haines, J.E.; Perez, E.; Munson, G.; Chen, J.; Kane, M.; McDonel, P.E.; Guttman, M.; Lander, E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016, 539, 452–455. [Google Scholar] [CrossRef]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Rackham, O.; Shearwood, A.-M.J.; Mercer, T.R.; Davies, S.M.; Mattick, J.S.; Filipovska, A. Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA 2011, 17, 2085–2093. [Google Scholar] [CrossRef] [Green Version]
- Bridges, M.C.; Daulagala, A.C.; Kourtidis, A. LNCcation: lncRNA localization and function. J. Cell Biol. 2021, 220, 202009045. [Google Scholar] [CrossRef] [PubMed]
- Noh, J.H.; Kim, K.M.; McClusky, W.G.; Abdelmohsen, K.; Gorospe, M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip. Rev. RNA 2018, 9, e1471. [Google Scholar] [CrossRef] [PubMed]
- Guh, C.-Y.; Hsieh, Y.-H.; Chu, H.-P. Functions and properties of nuclear lncRNAs-from systematically mapping the interactomes of lncRNAs. J. Biomed. Sci. 2020, 27, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, H.; Li, F.; Heindl, L.M.; He, X.; Yu, J.; Yang, J.; Ge, S.; Ruan, J.; Jia, R.; et al. Long Non-coding RNA LINC-PINT Suppresses Cell Proliferation and Migration of Melanoma via Recruiting EZH2. Front. Cell Dev. Biol. 2019, 7, 350. [Google Scholar] [CrossRef] [PubMed]
- Ju, C.; Liu, R.; Zhang, Y.-W.; Zhang, Y.; Zhou, R.; Sun, J.; Lv, X.-B.; Zhang, Z. Mesenchymal stem cell-associated lncRNA in osteogenic differentiation. Biomed. Pharmacother. 2019, 115, 108912. [Google Scholar] [CrossRef]
- Tang, Y.; He, Y.; Zhang, P.; Wang, J.; Fan, C.; Yang, L.; Xiong, F.; Zhang, S.; Gong, Z.; Nie, S.; et al. lncRNAs regulate the cytoskeleton and related Rho/ROCK signaling in cancer metastasis. Mol. Cancer 2018, 17, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Peng, W.-X.; Koirala, P.; Mo, Y.-Y. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017, 36, 5661–5667. [Google Scholar] [CrossRef]
- García-Padilla, C.; Lozano-Velasco, E.; López-Sánchez, C.; Garcia-Martínez, V.; Aranega, A.; Franco, D. Non-Coding RNAs in Retinoic Acid as Differentiation and Disease Drivers. Non-Coding RNA 2021, 7, 13. [Google Scholar] [CrossRef]
- Rotty, J.D.; Bear, J.E. Competition and collaboration between different actin assembly pathways allows for homeostatic control of the actin cytoskeleton. BioArchitecture 2015, 5, 27–34. [Google Scholar] [CrossRef] [Green Version]
- Hohmann, T.; Dehghani, F. The Cytoskeleton—A Complex Interacting Meshwork. Cells 2019, 8, 362. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Lee, P.; Wu, X. Molecular and cytoskeletal regulations in epidermal development. Semin. Cell Dev. Biol. 2017, 69, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Stradal, T.E.B.; Pusch, R.; Kliche, S. Molecular Regulation of Cytoskeletal Rearrangements During T Cell Signalling. Results Probl. Cell Differ. 2006, 43, 219–244. [Google Scholar] [CrossRef] [PubMed]
- Lens, S.M.A.; Medema, R.H. Cytokinesis defects and cancer. Nat. Rev. Cancer 2018, 19, 32–45. [Google Scholar] [CrossRef]
- Seetharaman, S.; Etienne-Manneville, S. Cytoskeletal Crosstalk in Cell Migration. Trends Cell Biol. 2020, 30, 720–735. [Google Scholar] [CrossRef] [PubMed]
- Aseervatham, J. Cytoskeletal Remodeling in Cancer. Biology 2020, 9, 385. [Google Scholar] [CrossRef]
- Aillaud, M.; Schulte, L.N. Emerging Roles of Long Noncoding RNAs in the Cytoplasmic Milieu. Non-Coding RNA 2020, 6, 44. [Google Scholar] [CrossRef]
- Ma, X.; Dang, Y.; Shao, X.; Chen, X.; Wu, F.; Li, Y. Ubiquitination and Long Non-coding RNAs Regulate Actin Cytoskeleton Regulators in Cancer Progression. Int. J. Mol. Sci. 2019, 20, 2997. [Google Scholar] [CrossRef] [Green Version]
- García-Padilla, C.; Domínguez, J.N.; Lodde, V.; Munk, R.; Abdelmohsen, K.; Gorospe, M.; Jiménez-Sábado, V.; Ginel, A.; Hove-Madsen, L.; Aránega, A.E.; et al. Identification of atrial-enriched lncRNA Walras linked to cardiomyocyte cytoarchitecture and atrial fibrillation. FASEB J. 2021, 36, e22051. [Google Scholar] [CrossRef]
- Wen, Q.; Janmey, P.A. Polymer physics of the cytoskeleton. Curr. Opin. Solid State Mater. Sci. 2011, 15, 177–182. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Zhang, L.; Shen, H.; Hao, Q.; Fu, S.; Liu, X. Up-regulation of long non-coding RNA CYTOR induced by icariin promotes the viability and inhibits the apoptosis of chondrocytes. BMC Complement. Med. Ther. 2021, 21, 152. [Google Scholar] [CrossRef]
- Liang, J.; Wei, X.; Liu, Z.; Cao, D.; Tang, Y.; Zou, Z.; Zhou, C.; Lu, Y. Long noncoding RNA CYTOR in cancer: A TCGA data review. Clin. Chim. Acta 2018, 483, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Hinds, M.; Smits, C.; Fredericks-Short, R.; Risk, J.M.; Bailey, M.; Huang, D.; Day, C. Bim, Bad and Bmf: Intrinsically unstructured BH3-only proteins that undergo a localized conformational change upon binding to prosurvival Bcl-2 targets. Cell Death Differ. 2007, 14, 128–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nallanthighal, S.; Heiserman, J.P.; Cheon, D.-J. The Role of the Extracellular Matrix in Cancer Stemness. Front. Cell Dev. Biol. 2019, 7, 86. [Google Scholar] [CrossRef] [PubMed]
- Martínez, P.T.; Navajas, P.L.; Lietha, D. FAK Structure and Regulation by Membrane Interactions and Force in Focal Adhesions. Biomolecules 2020, 10, 179. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Z.; Wei, W. RAB5A promotes the formation of filopodia in pancreatic cancer cells via the activation of cdc42 and β1-integrin. Biochem. Biophys. Res. Commun. 2021, 535, 54–59. [Google Scholar] [CrossRef]
- Du, D.-S.; Yang, X.-Z.; Wang, Q.; Dai, W.-J.; Kuai, W.-X.; Liu, Y.-L.; Chu, D.; Tang, X.-J. Effects of CDC42 on the proliferation and invasion of gastric cancer cells. Mol. Med. Rep. 2016, 13, 550–554. [Google Scholar] [CrossRef]
- Newell-Litwa, K.A.; Badoual, M.; Asmussen, H.; Patel, H.; Whitmore, L.; Horwitz, A.R. ROCK1 and 2 differentially regulate actomyosin organization to drive cell and synaptic polarity. J. Cell Biol. 2015, 210, 225–242. [Google Scholar] [CrossRef] [Green Version]
- Zanin-Zhorov, A.; Blazar, B.R. ROCK2, a critical regulator of immune modulation and fibrosis has emerged as a therapeutic target in chronic graft-versus-host disease. Clin. Immunol. 2021, 230, 108823. [Google Scholar] [CrossRef]
- Julian, L.; Olson, M.F. Rho-associated coiled-coil containing kinases (ROCK): Structure, regulation, and functions. Small GTPases 2014, 5, e29846. [Google Scholar] [CrossRef]
- Parri, M.; Chiarugi, P. Rac and Rho GTPases in cancer cell motility control. Cell Commun. Signal. 2010, 8, 23. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.; He, Y.; Shi, L.; Yang, L.; Wang, J.; Lian, Y.; Fan, C.; Zhang, P.; Guo, C.; Zhang, S.; et al. Co-expression of AFAP1-AS1 and PD-1 predicts poor prognosis in nasopharyngeal carcinoma. Oncotarget 2017, 8, 39001–39011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, T.; Advani, T.H.T.A.A. Inflammation in Cardiovascular Disease and Regulation of the Actin Cytoskeleton in Inflammatory Cells: The Actin Cytoskeleton as a Target. Cardiovasc. Hematol. Agents Med. Chem. 2006, 4, 165–182. [Google Scholar] [CrossRef] [PubMed]
- Caporizzo, M.A.; Chen, C.Y.; Prosser, B.L. Cardiac microtubules in health and heart disease. Exp. Biol. Med. 2019, 244, 1255–1272. [Google Scholar] [CrossRef] [PubMed]
- Eira, J.; Silva, C.S.; Sousa, M.; Liz, M.A. The cytoskeleton as a novel therapeutic target for old neurodegenerative disorders. Prog. Neurobiol. 2016, 141, 61–82. [Google Scholar] [CrossRef] [PubMed]
- Zatloukal, K.; Stumptner, C.; Fuchsbichler, A.; Fickert, P.; Lackner, C.; Trauner, M.; Denk, H. The keratin cytoskeleton in liver diseases. J. Pathol. 2004, 204, 367–376. [Google Scholar] [CrossRef]
- Akiyama, T.; Kawasaki, Y. Wnt signalling and the actin cytoskeleton. Oncogene 2006, 25, 7538–7544. [Google Scholar] [CrossRef] [Green Version]
- Galli, C.; Piemontese, M.; Lumetti, S.; Ravanetti, F.; Macaluso, G.; Passeri, G. Actin cytoskeleton controls activation of Wnt/β-catenin signaling in mesenchymal cells on implant surfaces with different topographies. Acta Biomater. 2012, 8, 2963–2968. [Google Scholar] [CrossRef]
- Wang, Q.; Symes, A.J.; Kane, C.A.; Freeman, A.; Nariculam, J.; Munson, P.; Thrasivoulou, C.; Masters, J.R.W.; Ahmed, A. A Novel Role for Wnt/Ca2+ Signaling in Actin Cytoskeleton Remodeling and Cell Motility in Prostate Cancer. PLoS ONE 2010, 5, e10456. [Google Scholar] [CrossRef]
- Lorenzon, A.; Calore, M.; Poloni, G.; De Windt, L.J.; Braghetta, P.; Rampazzo, A. Wnt/β-catenin pathway in arrhythmogenic cardiomyopathy. Oncotarget 2017, 8, 60640–60655. [Google Scholar] [CrossRef] [Green Version]
- Lozano-Velasco, E.; Garcia-Padilla, C.; Aránega, A.E.; Franco, D. Genetics of Atrial Fibrilation: In Search of Novel Therapeutic Targets. Cardiovasc. Hematol. Disord. Drug Targets 2019, 19, 183–194. [Google Scholar] [CrossRef]
- Su, I.H.; Dobenecker, M.W.; Dickinson, E.; Oser, M.; Basavaraj, A.; Marqueron, R.; Viale, A.; Reinberg, D.; Wülfing, C.; Tarakhovsky, A. Polycomb Group Protein Ezh2 Controls Actin Polymerization and Cell Signaling. Cell 2005, 121, 425–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, R.; Kong, P.; Zhang, F.; Shu, Y.-N.; Nie, X.; Dong, L.-H.; Lin, Y.-L.; Xie, X.-L.; Zhao, L.-L.; Zhang, X.-J.; et al. EZH2-mediated α-actin methylation needs lncRNA TUG1, and promotes the cortex cytoskeleton formation in VSMCs. Gene 2017, 616, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Sept, D.; Xu, J.; Pollard, T.D.; McCammon, J.A. Annealing Accounts for the Length of Actin Filaments Formed by Spontaneous Polymerization. Biophys. J. 1999, 77, 2911–2919. [Google Scholar] [CrossRef] [Green Version]
- Chánez-Paredes, S.; Montoya-García, A.; Schnoor, M. Cellular and pathophysiological consequences of Arp2/3 complex inhibition: Role of inhibitory proteins and pharmacological compounds. Cell. Mol. Life Sci. 2019, 76, 3349–3361. [Google Scholar] [CrossRef] [PubMed]
- Pollard, T.D.; Blanchoin, L.; Mullins, R.D. Molecular Mechanisms Controlling Actin Filament Dynamics in Nonmuscle Cells. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 545–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biber, G.; Ben-Shmuel, A.; Sabag, B.; Barda-Saad, M. Actin regulators in cancer progression and metastases: From structure and function to cytoskeletal dynamics. International Review of Cell and Molecular Biology 2020, 356, 131–196. [Google Scholar] [CrossRef]
- Yilmaz, M.; Christofori, G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009, 28, 15–33. [Google Scholar] [CrossRef] [Green Version]
- Jones, M.C.; Zha, J.; Humphries, M.J. Connections between the cell cycle, cell adhesion and the cytoskeleton. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180227. [Google Scholar] [CrossRef]
- Pollard, T.D. Actin and Actin-Binding Proteins. Cold Spring Harb. Perspect. Biol. 2016, 8, a018226. [Google Scholar] [CrossRef] [Green Version]
- Uray, K.; Major, E.; Lontay, B. MicroRNA Regulatory Pathways in the Control of the Actin–Myosin Cytoskeleton. Cells 2020, 9, 1649. [Google Scholar] [CrossRef]
- Pei, H.; Hu, W.; Guo, Z.; Chen, H.; Ma, J.; Mao, W.; Li, B.; Wang, A.; Wan, J.; Zhang, J.; et al. Long Noncoding RNA CRYBG3 Blocks Cytokinesis by Directly Binding G-Actin. Cancer Res. 2018, 78, 4563–4572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, K.L.; Kabbarah, O.; Liang, M.-C.; Ivanova, E.; Anagnostou, V.; Wu, J.; Dhakal, S.; Wu, M.; Chen, S.; Feinberg, T.; et al. GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature 2009, 459, 1085–1090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bass-Zubek, A.E.; Godsel, L.M.; Delmar, M.; Green, K.J. Plakophilins: Multifunctional scaffolds for adhesion and signaling. Curr. Opin. Cell Biol. 2009, 21, 708–716. [Google Scholar] [CrossRef] [Green Version]
- Van Grembergen, O.; Bizet, M.; de Bony, E.J.; Calonne, E.; Putmans, P.; Brohée, S.; Olsen, C.; Guo, M.; Bontempi, G.; Sotiriou, C.; et al. Portraying breast cancers with long noncoding RNAs. Sci. Adv. 2016, 2, e1600220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Jin, W.; Ma, D.; Cao, J.; Fu, T.; Zhang, Z.; Zhang, Y. Long non-coding RNA CYTOR regulates proliferation and metastasis of colon cancer cells through regulating miRNA-105/PTEN axis. Int. J. Clin. Experi Ment. Pathol. 2021, 14, 434–443. [Google Scholar]
- Tian, Q.; Yan, X.; Yang, L.; Liu, Z.; Yuan, Z.; Zhang, Y. lncRNA CYTOR promotes cell proliferation and tumor growth via miR-125b/SEMA4C axis in hepatocellular carcinoma. Oncol. Lett. 2021, 22, 1–12. [Google Scholar] [CrossRef]
- Wei, F.; Wang, Y.; Zhou, Y.; Li, Y. Long noncoding RNA CYTOR triggers gastric cancer progression by targeting miR-103/RAB10. Acta Biochim. et Biophys. Sin. 2021, 53, 1044–1054. [Google Scholar] [CrossRef]
- Chen, W.; Du, M.; Hu, X.; Ma, H.; Zhang, E.; Wang, T.; Yin, L.; He, X.; Hu, Z. Long noncoding RNA cytoskeleton regulator RNA promotes cell invasion and metastasis by titrating miR-613 to regulate ANXA2 in nasopharyngeal carcinoma. Cancer Med. 2019, 9, 1209–1219. [Google Scholar] [CrossRef]
- Ostrowska, Z.; Moraczewska, J. Cofilin – a protein controlling dynamics of actin filaments. Postępy Higieny i Medycyny Doświadczalnej 2017, 71, 339–351. [Google Scholar] [CrossRef]
- Shishkin, S.; Eremina, L.; Pashintseva, N.; Kovalev, L.; Kovaleva, M. Cofilin-1 and Other ADF/Cofilin Superfamily Members in Human Malignant Cells. Int. J. Mol. Sci. 2016, 18, 10. [Google Scholar] [CrossRef] [Green Version]
- Howard, J.; Goh, C.Y.; Gorzel, K.W.; Higgins, M.; McCann, A. The potential role of cofilin-1 in promoting triple negative breast cancer (TNBC) metastasis via the extracellular vesicles (EVs). Transl. Oncol. 2021, 15, 101247. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Huang, Y.; Zhao, J.; Wu, L.; Qi, Q.; Liu, Y.; Li, G.; Li, J.; Liu, H.; Wu, H. Cofilin: A Promising Protein Implicated in Cancer Metastasis and Apoptosis. Front. Cell Dev. Biol. 2021, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tao, L.; Jin, F.; Gu, H.; Dai, X.; Ni, T.; Feng, J.; Ding, Y.; Xiao, W.; Qian, Y.; et al. Cofilin 1 induces the epithelial-mesenchymal transition of gastric cancer cells by promoting cytoskeletal rearrangement. Oncotarget 2017, 8, 39131–39142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mousavi, S.; Ng, O.; Saunders, J.H.; Acheson, A.G.; Parsons, S.L. Study of cofilin 1 gene expression in colorectal cancer. J. Gastrointest. Oncol. 2018, 9, 791–796. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, P.; Liu, J.; Zheng, J.; Liu, Y.; Chen, J.; Xue, Y. Gas5 Exerts Tumor-suppressive Functions in Human Glioma Cells by Targeting miR-222. Mol. Ther. 2015, 23, 1899–1911. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; He, D.; Fu, Y.; Zhang, R.; Guo, H.; Wang, Z.; Wang, Y.; Gao, T.; Wei, Y.; Guo, Y.; et al. A novel lncRNA ARST represses glioma progression by inhibiting ALDOA-mediated actin cytoskeleton integrity. J. Exp. Clin. Cancer Res. 2021, 40, 187. [Google Scholar] [CrossRef]
- Cartron, P.-F.; Loussouarn, D.; Campone, M.; Martin, S.A.; Vallette, F. Prognostic impact of the expression/phosphorylation of the BH3-only proteins of the BCL-2 family in glioblastoma multiforme. Cell Death Dis. 2012, 3, e421. [Google Scholar] [CrossRef] [Green Version]
- Scott, G.A.; McClelland, L.A.; Fricke, A.F.; Fender, A. Plexin C1, A Receptor for Semaphorin 7A, Inactivates Cofilin and Is a Potential Tumor Suppressor for Melanoma Progression. J. Investig. Dermatol. 2009, 129, 954–963. [Google Scholar] [CrossRef] [Green Version]
- Fuse, M.; Kojima, S.; Enokida, H.; Chiyomaru, T.; Yoshino, H.; Nohata, N.; Kinoshita, T.; Sakamoto, S.; Naya, Y.; Nakagawa, M.; et al. Tumor suppressive microRNAs (miR-222 and miR-31) regulate molecular pathways based on microRNA expression signature in prostate cancer. J. Hum. Genet. 2012, 57, 691–699. [Google Scholar] [CrossRef] [Green Version]
- Garofalo, M.; Quintavalle, C.; Romano, G.; Croce, C.M.; Condorelli, G. miR221/222 in Cancer: Their Role in Tumor Progression and Response to Therapy. Curr. Mol. Med. 2012, 12, 27–33. [Google Scholar] [CrossRef]
- Quintavalle, C.; Garofalo, M.; Zanca, C.; Romano, G.; Iaboni, M.; Caro, M.D.B.D.; Martinez-Montero, J.C.; Incoronato, M.; Nuovo, G.; Croce, C.M.; et al. miR-221/222 overexpession in human glioblastoma increases invasiveness by targeting the protein phosphate PTPμ. Oncogene 2011, 31, 858–868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theocharis, A.D.; Skandalis, S.S.; Gialeli, C.; Karamanos, N.K. Extracellular matrix structure. Adv. Drug Deliv. Rev. 2016, 97, 4–27. [Google Scholar] [CrossRef] [PubMed]
- Paluch, E.K.; Aspalter, I.M.; Sixt, M. Focal Adhesion–Independent Cell Migration. Annu. Rev. Cell Dev. Biol. 2016, 32, 469–490. [Google Scholar] [CrossRef]
- Shih, Y.-P.; Sun, P.; Wang, A.; Lo, S.H. Tensin1 positively regulates RhoA activity through its interaction with DLC1. Biochim. et Biophys. Acta 2015, 1853, 3258–3265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, J.M.; Rodriguez, Y.A.R.; Jeong, K.; Ahn, E.-Y.E.; Lim, S.-T.S. Targeting focal adhesion kinase in cancer cells and the tumor microenvironment. Exp. Mol. Med. 2020, 52, 877–886. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.-C.; Diermeier, S.D.; Yu, A.T.; Brine, L.D.; Russo, S.; Bhatia, S.; Alsudani, H.; Kostroff, K.; Bhuiya, T.; Brogi, E.; et al. MaTAR25 lncRNA regulates the Tensin1 gene to impact breast cancer progression. Nat. Commun. 2020, 11, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Kelm, R.J.; Lamba, G.S.; Levis, J.E.; Holmes, C.E. Characterization of purine-rich element binding protein B as a novel biomarker in acute myelogenous leukemia prognostication. J. Cell. Biochem. 2018, 119, 2073–2083. [Google Scholar] [CrossRef] [PubMed]
- Hall, E.H.; Daugherty, A.E.; Choi, C.K.; Horwitz, A.F.; Brautigan, D.L. Tensin1 Requires Protein Phosphatase-1α in Addition to RhoGAP DLC-1 to Control Cell Polarization, Migration, and Invasion. J. Biol. Chem. 2009, 284, 34713–34722. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Duncan, I.C.; Bozorgchami, H.; Lo, S.H. Tensin1 and a previously undocumented family member, tensin2, positively regulate cell migration. Proc. Natl. Acad. Sci. USA 2002, 99, 733–738. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Yi, Q.; Tang, L. The roles of nuclear focal adhesion kinase (FAK) on Cancer: A focused review. J. Exp. Clin. Cancer Res. 2019, 38, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Degirmenci, U.; Wang, M.; Hu, J. Targeting Aberrant RAS/RAF/MEK/ERK Signaling for Cancer Therapy. Cells 2020, 9, 198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Y.; Wei, G.; Zhang, L.; Zhou, H.; Wang, W.; Guo, P.; Cheng, C.; Ji, L.; Cai, Q.; Feng, Y.; et al. LncRNA DARS-AS1 aggravates the growth and metastasis of hepatocellular carcinoma via regulating the miR-3200-5p-Cytoskeleton associated protein 2 (CKAP2) axis. Bioeng. 2021, 12, 8217–8232. [Google Scholar] [CrossRef] [PubMed]
- Machesky, L.M. Lamellipodia and filopodia in metastasis and invasion. FEBS Lett. 2008, 582, 2102–2111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, M.; Pang, H.; Li, X.; Li, H.; Pan, J.; Chen, W. Long non-coding RNA urothelial cancer-associated 1 promotes bladder cancer cell migration and invasion by way of the hsa-miR-145– ZEB 1/2– FSCN 1 pathway. Cancer Sci. 2015, 107, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, Y.; Li, L.; Cao, J.; Guo, Y.; Wu, Y.; Gao, W. Fascin actin-bundling protein 1 in human cancer: Promising biomarker or therapeutic target? Mol. Ther. Oncolytics 2021, 20, 240–264. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, M. Involvement of partial EMT in cancer progression. J. Biochem. 2018, 164, 257–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanrahan, K.; O’Neill, A.; Prencipe, M.; Bugler, J.; Murphy, L.; Fabre, A.; Puhr, M.; Culig, Z.; Murphy, K.; Watson, R.W. The role of epithelial-mesenchymal transition drivers ZEB1 and ZEB2 in mediating docetaxel-resistant prostate cancer. Mol. Oncol. 2017, 11, 251–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burridge, K.; Wennerberg, K. Rho and Rac Take Center Stage. Cell 2004, 116, 167–179. [Google Scholar] [CrossRef] [Green Version]
- Amano, M.; Nakayama, M.; Kaibuchi, K. Rho-kinase/ROCK: A key regulator of the cytoskeleton and cell polarity. Cytoskeleton 2010, 67, 545–554. [Google Scholar] [CrossRef] [Green Version]
- Symons, M. Rho family GTPases: The cytoskeleton and beyond. Trends Biochem. Sci. 1996, 21, 178–181. [Google Scholar] [CrossRef]
- Jaffe, A.B.; Hall, A. RHO GTPASES: Biochemistry and Biology. Annu. Rev. Cell Dev. Biol. 2005, 21, 247–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svensmark, J.H.; Brakebusch, C. Rho GTPases in cancer: Friend or foe? Oncogene 2019, 38, 7447–7456. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.L.; Bement, W.M. Regulation of cytokinesis by Rho GTPase flux. Nat. Cell Biol. 2009, 11, 71–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joshi, B.; Strugnell, S.S.; Goetz, J.; Kojic, L.D.; Cox, M.E.; Griffith, O.; Chan, S.K.; Jones, S.; Leung, S.-P.; Masoudi, H.; et al. Phosphorylated Caveolin-1 Regulates Rho/ROCK-Dependent Focal Adhesion Dynamics and Tumor Cell Migration and Invasion. Cancer Res. 2008, 68, 8210–8220. [Google Scholar] [CrossRef] [Green Version]
- García-Mariscal, A.; Li, H.; Pedersen, E.; Peyrollier, K.; Ryan, K.M.; Stanley, A.; Quondamatteo, F.; Brakebusch, C. Loss of RhoA promotes skin tumor formation and invasion by upregulation of RhoB. Oncogene 2018, 37, 847–860. [Google Scholar] [CrossRef]
- Gallo, G. RhoA-kinase coordinates F-actin organization and myosin II activity during semaphorin-3A-induced axon retraction. J. Cell Sci. 2006, 119, 3413–3423. [Google Scholar] [CrossRef] [Green Version]
- Heasman, S.J.; Ridley, A.J. Multiple roles for RhoA during T cell transendothelial migration. Small GTPases 2010, 1, 174–179. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Borja, M.; Janssen, L.; Verwoerd, D.; Hordijk, P.; Neefjes, J. RhoB regulates endosome transport by promoting actin assembly on endosomal membranes through Dia1. J. Cell Sci. 2005, 118, 2661–2670. [Google Scholar] [CrossRef] [Green Version]
- Egami, Y.; Kawai, K.; Araki, N. RhoC regulates actin remodeling to form phagosomes during FcγR-mediated phagocytosis. J. Cell Sci. 2017, 130, 4168–4179. [Google Scholar] [CrossRef] [Green Version]
- Bompard, G.; Sharp, S.J.; Freiss, G.; Machesky, L. Involvement of Rac in actin cytoskeleton rearrangements induced by MIM-B. J. Cell Sci. 2005, 118, 5393–5403. [Google Scholar] [CrossRef] [Green Version]
- Kurokawa, K.; Itoh, R.; Yoshizaki, H.; Nakamura, Y.O.T.; Matsuda, M. Coactivation of Rac1 and Cdc42 at Lamellipodia and Membrane Ruffles Induced by Epidermal Growth Factor. Mol. Biol. Cell 2004, 15, 1003–1010. [Google Scholar] [CrossRef] [Green Version]
- Mehidi, A.; Rossier, O.; Schaks, M.; Chazeau, A.; Binamé, F.; Remorino, A.; Coppey, M.; Karatas, Z.; Sibarita, J.-B.; Rottner, K.; et al. Transient Activations of Rac1 at the Lamellipodium Tip Trigger Membrane Protrusion. Curr. Biol. 2019, 29, 2852–2866.e5. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, S.K.; Cabrera, R.; Mao, S.P.; Christin, J.R.; Wu, B.; Guo, W.; Bravo-Cordero, J.J.; Condeelis, J.S.; Segall, J.E.; Hodgson, L. Rac3 regulates breast cancer invasion and metastasis by controlling adhesion and matrix degradation. J. Cell Biol. 2017, 216, 4331–4349. [Google Scholar] [CrossRef] [PubMed]
- Masi, I.; Caprara, V.; Spadaro, F.; Chellini, L.; Sestito, R.; Zancla, A.; Rainer, A.; Bagnato, A.; Rosanò, L. Endothelin-1 drives invadopodia and interaction with mesothelial cells through ILK. Cell Rep. 2021, 34, 108800. [Google Scholar] [CrossRef] [PubMed]
- Arana, E.; Vehlow, A.; Harwood, N.E.; Vigorito, E.; Henderson, R.; Turner, M.; Tybulewicz, V.; Batista, F.D. Activation of the Small GTPase Rac2 via the B Cell Receptor Regulates B Cell Adhesion and Immunological-Synapse Formation. Immun. 2008, 28, 88–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krugmann, S.; Jordens, I.; Gevaert, K.; Driessens, M.; Vandekerckhove, J.; Hall, A. Cdc42 induces filopodia by promoting the formation of an IRSp53:Mena complex. Curr. Biol. 2001, 11, 1645–1655. [Google Scholar] [CrossRef] [Green Version]
- Aikemu, B.; Shao, Y.; Yang, G.; Ma, J.; Zhang, S.; Yang, X.; Hong, H.; Yesseyeva, G.; Huang, L.; Jia, H.; et al. NDRG1 regulates Filopodia-induced Colorectal Cancer invasiveness via modulating CDC42 activity. Int. J. Biol. Sci. 2021, 17, 1716–1730. [Google Scholar] [CrossRef]
- Xiao, X.-H.; Lv, L.-C.; Duan, J.; Wu, Y.-M.; He, S.-J.; Hu, Z.-Z.; Xiong, L.-X. Regulating Cdc42 and Its Signaling Pathways in Cancer: Small Molecules and MicroRNA as New Treatment Candidates. Molecules 2018, 23, 787. [Google Scholar] [CrossRef] [Green Version]
- Jerrell, R.J.; Leih, M.J.; Parekh, A. The ROCK isoforms differentially regulate the morphological characteristics of carcinoma cells. Small GTPases 2020, 11, 131–137. [Google Scholar] [CrossRef] [Green Version]
- Darenfed, H.; Dayanandan, B.; Zhang, T.; Hsieh, S.H.-K.; Fournier, A.E.; Mandato, C.A. Molecular characterization of the effects of Y-27632. Cell Motil. Cytoskelet. 2007, 64, 97–109. [Google Scholar] [CrossRef]
- Chin, V.T.; Nagrial, A.M.; Chou, A.; Biankin, A.V.; Gill, A.J.; Timpson, P.; Pajic, M. Rho-associated kinase signalling and the cancer microenvironment: Novel biological implications and therapeutic opportunities. Expert Rev. Mol. Med. 2015, 17, e17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, C.-M.; Wei, L.; Au, S.L.-K.; Fan, D.N.-Y.; Zhou, Y.; Tsang, F.H.-C.; Law, C.-T.; Lee, J.M.-F.; He, X.; Shi, J.; et al. MiR-200b/200c/429 subfamily negatively regulates Rho/ROCK signaling pathway to suppress hepatocellular carcinoma metastasis. Oncotarget 2015, 6, 13658–13670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, X.; Liu, Y.; Yang, W.; Xia, Y.; Yang, C.; Yang, S.; Liu, X. Long noncoding RNA MALAT1 as a potential therapeutic target in osteosarcoma. J. Orthop. Res. 2016, 34, 932–941. [Google Scholar] [CrossRef] [Green Version]
- Chou, J.; Wang, B.; Zheng, T.; Li, X.; Zheng, L.; Hu, J.; Zhang, Y.; Xing, Y.; Xi, T. MALAT1 induced migration and invasion of human breast cancer cells by competitively binding miR-1 with cdc42. Biochem. Biophys. Res. Commun. 2016, 472, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Rozenchan, P.B.; Pasini, F.S.; Roela, R.A.; Katayama, M.L.H.; Mundim, F.G.L.; Brentani, H.; Lyra, E.C.; Brentani, M.M. Specific upregulation of RHOA and RAC1 in cancer-associated fibroblasts found at primary tumor and lymph node metastatic sites in breast cancer. Tumor Biol. 2015, 36, 9589–9597. [Google Scholar] [CrossRef]
- Ge, Z.; Cheng, Z.; Yang, X.; Huo, X.; Wang, N.; Wang, H.; Wang, C.; Gu, D.; Zhao, F.; Yao, M.; et al. Long noncoding RNASchLAHsuppresses metastasis of hepatocellular carcinoma through interacting with fused in sarcoma. Cancer Sci. 2017, 108, 653–662. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Yan, G.; Zhang, Y.; Jia, X.; Bu, P. Long non-coding RNA MEG3 suppresses migration and invasion of thyroid carcinoma by targeting of Rac1. Neoplasma 2015, 62, 541–549. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Wu, X.; Surma, M.; Vemula, S.; Zhang, L.; Yang, Y.; Kapur, R.; Wei, L. Distinct roles for ROCK1 and ROCK2 in the regulation of cell detachment. Cell Death Dis. 2013, 4, e483. [Google Scholar] [CrossRef] [Green Version]
- Shen, D.; Liu, Y.; Liu, Y.; Wang, T.; Yuan, L.; Huang, X.; Wang, Y. Long non-coding RNA EWSAT1 promoted metastasis and actin cytoskeleton changes via miR-24-3p sponging in osteosarcoma. J. Cell. Mol. Med. 2021, 25, 716–728. [Google Scholar] [CrossRef]




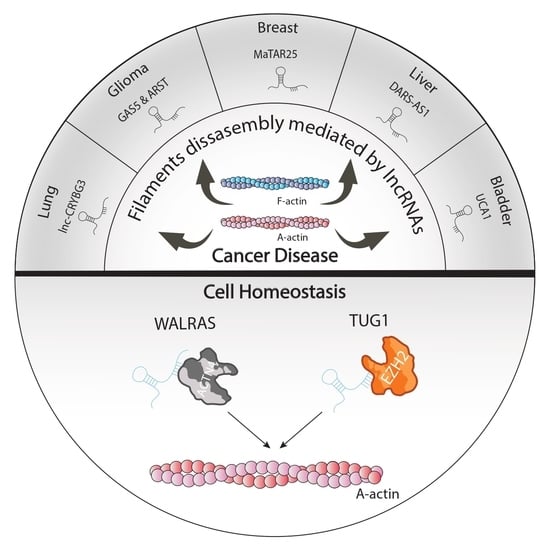
| Cytoskeletal-lncRNAs Related to Tumorigenesis | ||||
|---|---|---|---|---|
| lncRNA | Target Molecule | Function | Tissue or Cell Line | Reference |
| LNC-CRYBG3 | G-actin | Inhibition of F-actin polymerization avoiding G-actin phosphorylation | Lung cancer | [28] |
| CYTOR | Golph3, Rhobtb3 and PKP4 | Cytoskeletal homeostasis and cell cycle progression | Breast cancer cell line | [29] |
| Gas5 | miR-222 | Enhance Bmf and PLXN1 expression reducing aggressiveness tumour | U87 and U251 glioma cell line | [30] |
| ARST | ALDOA | Mediate actin fibers integrity avoiding that ALDOA can attach to F-actin binding sites increasing F-actin depolymeration | U87 and U251 glioma cell line | [31] |
| MaTaR25 | PURB and Tensin1 | Enhance PURB dependent genes remodelling cytoskeleton architecture and increasing migration and spread out of maligned cells | Breast cancer cell line | [32] |
| DARS-AS1 | miR-3002 | Sponge miR-3002 enhancing CKAP2 translation and aggravating the growth and metastasis of tumor | Hepatocellular carcinoma | [33] |
| UCA1 | ZEB1/2 and FSCN1 | Increase formation of actin-dependent cell filopodia enhancing metastasis | Bladder carcinoma | [34] |
| Malat1 | RhoA, ROCK1 and ROCK2 | Increasing RhoA, ROCK1, and ROCK2 translation required to migration and cytoskeletal homeostasis | Osteosarcoma | [35] |
| Malat1 | miR-1 | Sponge miR-1 enhancing Cdc42 translation required to migration and cytoskeletal homeostasis | Breast carcinoma | [36] |
| AFAP1-AS1 | RhoA and Rac1 | Enhancing progression and poor prognosis of nasopharyngeal carcinoma increasing capacity of spreading out | nasopharyngeal carcinoma | [37] |
| SchLAH | FUS/TLS | Repressing cellular migration and therein metastasis triggering downregulation of RhoA/Rac2 signalling | Lung carcinoma | [38] |
| EWAST1 | miR-24-3p | Sponge miR-24-3p enhancing expression of ROCK1 and promoting actin stress fiber formation and migration | Osteosarcoma | [39] |
| Walras | ACTN4 | Required to actin cytoskeleton integrity | Cardiomyocites | [40] |
| TUG1 | EZH2 and actin | Methylation of α-actin by EZH2 | Vascular smooth muscle cells | [41] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Padilla, C.; Muñoz-Gallardo, M.d.M.; Lozano-Velasco, E.; Castillo-Casas, J.M.; Caño-Carrillo, S.; García-López, V.; Aránega, A.; Franco, D.; García-Martínez, V.; López-Sánchez, C. New Insights into the Roles of lncRNAs as Modulators of Cytoskeleton Architecture and Their Implications in Cellular Homeostasis and in Tumorigenesis. Non-Coding RNA 2022, 8, 28. https://doi.org/10.3390/ncrna8020028
García-Padilla C, Muñoz-Gallardo MdM, Lozano-Velasco E, Castillo-Casas JM, Caño-Carrillo S, García-López V, Aránega A, Franco D, García-Martínez V, López-Sánchez C. New Insights into the Roles of lncRNAs as Modulators of Cytoskeleton Architecture and Their Implications in Cellular Homeostasis and in Tumorigenesis. Non-Coding RNA. 2022; 8(2):28. https://doi.org/10.3390/ncrna8020028
Chicago/Turabian StyleGarcía-Padilla, Carlos, María del Mar Muñoz-Gallardo, Estefanía Lozano-Velasco, Juan Manuel Castillo-Casas, Sheila Caño-Carrillo, Virginio García-López, Amelia Aránega, Diego Franco, Virginio García-Martínez, and Carmen López-Sánchez. 2022. "New Insights into the Roles of lncRNAs as Modulators of Cytoskeleton Architecture and Their Implications in Cellular Homeostasis and in Tumorigenesis" Non-Coding RNA 8, no. 2: 28. https://doi.org/10.3390/ncrna8020028
APA StyleGarcía-Padilla, C., Muñoz-Gallardo, M. d. M., Lozano-Velasco, E., Castillo-Casas, J. M., Caño-Carrillo, S., García-López, V., Aránega, A., Franco, D., García-Martínez, V., & López-Sánchez, C. (2022). New Insights into the Roles of lncRNAs as Modulators of Cytoskeleton Architecture and Their Implications in Cellular Homeostasis and in Tumorigenesis. Non-Coding RNA, 8(2), 28. https://doi.org/10.3390/ncrna8020028