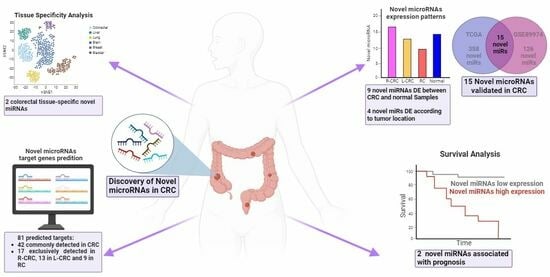
Discovery of Novel miRNAs in Colorectal Cancer: Potential Biological Roles and Clinical Utility
Abstract
:1. Introduction
2. Results
2.1. Identification of Novel microRNAs
In Silico Validation of the Novel miRNAs
2.2. Genomic Location and Tissue-Specific Expression Patterns of Novel miRNAs
2.2.1. Genomic Location
2.2.2. Tissue-Specific Expression Patterns of Novel miRNAs
2.3. Prognostic Relevance
2.4. Target Prediction and Expression in CRC
2.5. Interaction Networks
3. Discussion
4. Materials and Methods
4.1. Study Cohorts
4.2. Small-RNA Sequencing Processing and miRNA Discovery
4.3. Novel microRNA Analysis
4.4. Gene Network
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- INCA (Instituto Nacional de Câncer José Alencar Gomes da Silva). Incidência de Câncer No Brasil 2023; Neoplasia Maligna Do Cólon e Reto (Taxas Ajustadas): Português, Brasil, 2022. [Google Scholar]
- The Cancer Genome Atlas Network. Comprehensive Molecular Characterization of Human Colon and Rectal Cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef]
- Palazzo, A.F.; Lee, E.S. Non-Coding RNA: What Is Functional and What Is Junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An Overview of MicroRNAs: Biology, Functions, Therapeutics, and Analysis Methods. J. Cell Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Yang, J.B.; Lin, X.Y.; Xie, B.L.; Xu, Y.X.; Lin, S.; Xu, T.W. Recent Advances in the Diagnostic and Therapeutic Roles of MicroRNAs in Colorectal Cancer Progression and Metastasis. Front. Oncol. 2022, 12, 911856. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Lizano, E.; Houben, A.J.S.; Bezdan, D.; Báñez-Coronel, M.; Kudla, G.; Mateu-Huertas, E.; Kagerbauer, B.; González, J.; Chen, K.C.; et al. Evidence for the Biogenesis of More than 1000 Novel Human MicroRNAs. Genome Biol. 2014, 15, R57. [Google Scholar] [CrossRef]
- Londin, E.; Loher, P.; Telonis, A.G.; Quann, K.; Clark, P.; Jing, Y.; Hatzimichael, E.; Kirino, Y.; Honda, S.; Lally, M.; et al. Analysis of 13 Cell Types Reveals Evidence for the Expression of Numerous Novel Primate- and Tissue-Specific MicroRNAs. Proc. Natl. Acad. Sci. USA 2015, 112, E1106–E1115. [Google Scholar] [CrossRef]
- Rock, L.D.; Minatel, B.C.; Marshall, E.A.; Guisier, F.; Sage, A.P.; Camargo Barros-Filho, M.; Stewart, G.L.; Garnis, C.; Lam, W.L. Expanding the Transcriptome of Head and Neck Squamous Cell Carcinoma Through Novel MicroRNA Discovery. Front. Oncol. 2019, 9, 1305. [Google Scholar] [CrossRef]
- Pewarchuk, M.E.; Barros-Filho, M.C.; Minatel, B.C.; Cohn, D.E.; Guisier, F.; Sage, A.P.; Marshall, E.A.; Stewart, G.L.; Rock, L.D.; Garnis, C.; et al. Upgrading the Repertoire of MiRNAs in Gastric Adenocarcinoma to Provide a New Resource for Biomarker Discovery. Int. J. Mol. Sci. 2019, 20, 5697. [Google Scholar] [CrossRef]
- Barros-Filho, M.C.; Pewarchuk, M.; Minatel, B.d.C.; Sage, A.P.; Marshall, E.A.; Martinez, V.D.; Rock, L.D.; MacAulay, G.; Kowalski, L.P.; Rogatto, S.R.; et al. Previously Undescribed Thyroid-Specific MiRNA Sequences in Papillary Thyroid Carcinoma. J. Hum. Genet. 2019, 64, 505–508. [Google Scholar] [CrossRef] [PubMed]
- Reshmi, G.; Chandra, S.S.V.; Babu, V.J.M.; Babu, P.S.S.; Santhi, W.S.; Ramachandran, S.; Lakshmi, S.; Nair, A.S.; Pillai, M.R. Identification and Analysis of Novel MicroRNAs from Fragile Sites of Human Cervical Cancer: Computational and Experimental Approach. Genomics 2011, 97, 333–340. [Google Scholar] [CrossRef]
- Sage, A.P.; Minatel, B.C.; Marshall, E.A.; Martinez, V.D.; Stewart, G.L.; Enfield, K.S.S.S.; Lam, W.L. Expanding the MiRNA Transcriptome of Human Kidney and Renal Cell Carcinoma. Int. J. Genomics 2018, 2018, 6972397. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, Z.; Mal, C. Functional Role of Hub Molecules in MiRNA and Transcription Factor Mediated Gene Regulatory Network of Colorectal and Lung Cancer. Gene Rep. 2021, 23, 101129. [Google Scholar] [CrossRef]
- Mirceta, M.; Shum, N.; Schmidt, M.H.M.; Pearson, C.E. Fragile Sites, Chromosomal Lesions, Tandem Repeats, and Disease. Front. Genet. 2022, 13, 985975. [Google Scholar] [CrossRef] [PubMed]
- Debacker, K.; Frank Kooy, R. Fragile Sites and Human Disease. Hum. Mol. Genet. 2007, 16, R150–R158. [Google Scholar] [CrossRef]
- Laganà, A.; Russo, F.; Sismeiro, C.; Giugno, R.; Pulvirenti, A.; Ferro, A. Variability in the Incidence of MiRNAs and Genes in Fragile Sites and the Role of Repeats and CpG Islands in the Distribution of Genetic Material. PLoS ONE 2010, 5, e11166. [Google Scholar] [CrossRef]
- Szczepanek, J.; Skorupa, M.; Tretyn, A. MicroRNA as a Potential Therapeutic Molecule in Cancer. Cells 2022, 11, 1008. [Google Scholar] [CrossRef]
- Thomas, J.; Ohtsuka, M.; Pichler, M.; Ling, H. MicroRNAs: Clinical Relevance in Colorectal Cancer. Int. J. Mol. Sci. 2015, 16, 28063–28076. [Google Scholar] [CrossRef]
- Slattery, M.L.; Wolff, E.; Hoffman, M.D.; Pellatt, D.F.; Milash, B.; Wolff, R.K. MicroRNAs and Colon and Rectal Cancer: Differential Expression by Tumor Location and Subtype. Genes. Chromosomes Cancer 2011, 50, 196–206. [Google Scholar] [CrossRef]
- Omrane, I.; Kourda, N.; Stambouli, N.; Privat, M.; Medimegh, I.; Arfaoui, A.; Uhrhammer, N.; Bougatef, K.; Baroudi, O.; Bouzaienne, H.; et al. MicroRNAs 146a and 147b Biomarkers for Colorectal Tumor’s Localization. Biomed. Res. Int. 2014, 2014, 584852. [Google Scholar] [CrossRef]
- Coebergh Van Den Braak, R.R.J.; Sieuwerts, A.M.; Lalmahomed, Z.S.; Smid, M.; Wilting, S.M.; Bril, S.I.; Xiang, S.; Van Der Vlugt-Daane, M.; De Weerd, V.; Van Galen, A.; et al. Confirmation of a Metastasis-Specific MicroRNA Signature in Primary Colon Cancer. Sci. Rep. 2018, 8, 5242. [Google Scholar] [CrossRef]
- Yang, L.; Li, L.; Ma, J.; Yang, S.; Zou, C.; Yu, X. MiRNA and MRNA Integration Network Construction Reveals Novel Key Regulators in Left-Sided and Right-Sided Colon Adenocarcinoma. Biomed. Res. Int. 2019, 2019, 7149296. [Google Scholar] [CrossRef]
- Eneh, S.; Heikkinen, S.; Hartikainen, J.M.; Kuopio, T.; Mecklin, J.P.; Kosma, V.M.; Mannermaa, A. MicroRNAs Associated with Biological Pathways of Left- And Right-Sided Colorectal Cancer. Anticancer. Res. 2020, 40, 3713–3722. [Google Scholar] [CrossRef]
- Loree, J.M.; Pereira, A.A.L.; Lam, M.; Willauer, A.N.; Raghav, K.; Dasari, A.; Van Morris, K.; Advani, S.; Menter, D.G.; Eng, C.; et al. Classifying Colorectal Cancer by Tumor Location Rather than Sidedness Highlights a Continuum in Mutation Profiles and Consensus Molecular Subtypes. Clin. Cancer Res. 2018, 24, 1062–1072. [Google Scholar] [CrossRef]
- Dai, Y.; Duan, H.; Duan, C.; Zhu, H.; Zhou, R.; Pei, H.; Shen, L. TCF21 Functions as a Tumor Suppressor in Colorectal Cancer through Inactivation of PI3K/AKT Signaling. Onco Targets Ther. 2017, 10, 1603–1611. [Google Scholar] [CrossRef] [PubMed]
- Noetzel, E.; Rose, M.; Sevinc, E.; Hilgers, R.D.; Hartmann, A.; Naami, A.; Knüchel, R.; Dahl, E. Intermediate Filament Dynamics and Breast Cancer: Aberrant Promoter Methylation of the Synemin Gene Is Associated with Early Tumor Relapse. Oncogene 2010, 29, 4814–4825. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Y.; Xiao, S.; Fu, K. Integrated Analysis of the Functions and Prognostic Values of RNA-Binding Proteins in Colorectal Cancer. Front. Cell Dev. Biol. 2020, 8, 595605. [Google Scholar] [CrossRef]
- Lai, S.H.; Zervoudakis, G.; Chou, J.; Gurney, M.E.; Quesnelle, K.M. PDE4 Subtypes in Cancer. Oncogene 2020, 39, 3791. [Google Scholar] [CrossRef]
- Chen, L.; Gao, H.; Liang, J.; Qiao, J.; Duan, J.; Shi, H.; Zhen, T.; Li, H.; Zhang, F.; Zhu, Z.; et al. MiR-203a-3p Promotes Colorectal Cancer Proliferation and Migration by Targeting PDE4D. Am. J. Cancer Res. 2018, 8, 2387. [Google Scholar]
- Chung, Y.; Kim, H.; Bang, S.; Jang, K.; Paik, S.S.; Shin, S.-J. Nuclear Expression Loss of SSBP2 Is Associated with Poor Prognostic Factors in Colorectal Adenocarcinoma. Diagnostics 2020, 10, 1097. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Klumpp, S.; Amin, H.M.; Liang, H.; Li, J.; Estrov, Z.; Zweidler-McKay, P.; Brandt, S.J.; Agulnick, A.; Nagarajan, L. SSBP2 Is an in Vivo Tumor Suppressor and Regulator of LDB1 Stability. Oncogene 2010, 29, 3044. [Google Scholar] [CrossRef]
- Zhu, X.; Shen, Z.; Man, D.; Ruan, H.; Huang, S. MiR-152-3p Affects the Progression of Colon Cancer via the KLF4/IFITM3 Axis. Comput. Math. Methods Med. 2020, 2020, 8209504. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Cheung, J.; Bialkowska, A.B. Krüppel-like Factors 4 and 5 in Colorectal Tumorigenesis. Cancers 2023, 15, 2430. [Google Scholar] [CrossRef] [PubMed]
- Chang, G.; Xu, S.; Dhir, R.; Chandran, U.; O’Keefe, D.S.; Greenberg, N.M.; Gingrich, J.R. Hypoexpression and Epigenetic Regulation of Candidate Tumor Suppressor Gene CADM-2 in Human Prostate Cancer. Clin. Cancer Res. 2010, 16, 5390. [Google Scholar] [CrossRef]
- Semba, S.; Itoh, N.; Ito, M.; Youssef, E.M.; Harada, M.; Moriya, T.; Kimura, W.; Yamakawa, M. Down-Regulation of PIK3CG, a Catalytic Subunit of Phosphatidylinositol 3-OH Kinase, by CpG Hypermethylation in Human Colorectal Carcinoma. Clin. Cancer Res. 2002, 8, 3824–3831. [Google Scholar] [PubMed]
- Wang, Y.; Zhao, J.; Wang, Y.; Gao, J.; Yang, H.; Li, H. MiR-17-5p Targets and Downregulates CADM2, Activating the Malignant Phenotypes of Colon Cancer Cells. Mol. Biotechnol. 2022, 64, 1388–1400. [Google Scholar] [CrossRef]
- Cui, X.; Liang, H.; Hao, C.; Jing, X. Homer1 Is a Potential Biomarker for Prognosis in Human Colorectal Carcinoma, Possibly in Association with G3BP1 Signaling. Cancer Manag. Res. 2020, 12, 2899. [Google Scholar] [CrossRef]
- Arribas, J.; Cajuso, T.; Rodio, A.; Marcos, R.; Leonardi, A.; Velázquez, A. NF-ΚB Mediates the Expression of TBX15 in Cancer Cells. PLoS ONE 2016, 11, e0157761. [Google Scholar] [CrossRef]
- Martinez, V.D.; Marshall, E.A.; Anderson, C.; Ng, K.W.; Minatel, B.C.; Sage, A.P.; Enfield, K.S.S.; Xu, Z.; Lam, W.L. Discovery of Previously Undetected MicroRNAs in Mesothelioma and Their Use as Tissue-of-Origin Markers. Am. J. Respir. Cell Mol. Biol. 2019, 61, 266–268. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A Web Tool for Visualizing Clustering of Multivariate Data Using Principal Component Analysis and Heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef]
- Kumar, R.; Nagpal, G.; Kumar, V.; Usmani, S.S.; Agrawal, P.; Raghava, G.P.S. HumCFS: A Database of Fragile Sites in Human Chromosomes. BMC Genom. 2019, 19, 985. [Google Scholar] [CrossRef] [PubMed]
- Demšar, J.; Erjavec, A.; Hočevar, T.; Milutinovič, M.; Možina, M.; Toplak, M.; Umek, L.; Zbontar, J.; Zupan, B. Orange: Data Mining Toolbox in Python Tomaž Curk Matija Polajnar Laň Zagar. J. Mach. Learn. Res. 2013, 14, 2349–2353. [Google Scholar]
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, X. Prediction of Functional MicroRNA Targets by Integrative Modeling of MicroRNA Binding and Target Expression Data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. MiRDB: An Online Database for Prediction of Functional MicroRNA Targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and Interpreting Cancer Genomics Data via the Xena Platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING Database in 2023: Protein–Protein Association Networks and Functional Enrichment Analyses for Any Sequenced Genome of Interest. Nucleic Acids Res 2023, 51, D638. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498. [Google Scholar] [CrossRef]






| Novel miRNA | Discovery Group FC 1 | p-Value | Validation Group FC 1 | p-Value |
|---|---|---|---|---|
| nov-miR-13844-5p | - 2 | 4.15 × 10−31 | 2.4 | 1.83 × 10−1 |
| nov-miR-766-3p | 25.753 | 2.33 × 10−7 | 1.5 | 9.21 × 10−2 |
| nov-miR-7154-5p | 6.561 | 2.49 × 10−5 | 3.8 | 1.12 × 10−6 |
| nov-miR-13172-3p | 5.651 | 4.32 × 10−4 | 1.9 | 2.12 × 10−3 |
| nov-miR-3345-5p | 4.558 | 4.21 × 10−5 | 1.5 | 2.72 × 10−2 |
| nov-miR-2199-5p | 2.002 | 5.69 × 10−1 | 3.2 | 1.18 × 10−4 |
| nov-miR-8861-5p | 1.732 | 1.41 × 10−1 | 2.6 | 5.26 × 10−3 |
| nov-miR-5035-3p | 0.131 | 4.19 × 10−5 | 0.6 | 2.74 × 10−1 |
| nov-miR-5065-5p | 0.121 | 4.43 × 10−4 | 0.7 | 2.23 × 10−1 |
| nov-miR-1156-3p | 0.201 | 3.05 × 10−3 | 1.0 | 9.41 × 10−1 |
| nov-miR-8999-3p | 0.032 | 1.94 × 10−2 | 0.4 | 8.76 × 10−2 |
| nov-miR-1156-5p | 0.674 | 1.49 × 10−1 | 1.0 | 9.41 × 10−1 |
| nov-miR-651-5p | 0.671 | 7.35 × 10−1 | 0.9 | 2.70 × 10−1 |
| nov-miR-590-5p | 0.476 | 7.22 × 10−1 | 0.6 | 5.45 × 10−2 |
| nov-miR-13996-5p | 0.871 | 8.32 × 10−1 | 0.6 | 5.07 × 10−1 |
| nov-miR-9377-5p | 15.757 | 3.46 × 10−8 | 1.1 | 5.89 × 10−1 |
| nov-miR-6348-3p | 0.789 | 2.18 × 10−1 | 5.0 | 2.10 × 10−4 |
| nov-miR-13634-5p | 2.784 | 1.77 × 10−3 | 0.8 | 4.09 × 10−1 |
| nov-miR-171-3p | 0.450 | 4.80 × 10−3 | 3.0 | 8.55 × 10−2 |
| nov-miR-4285-5p | 0.172 | 3.42 × 10−8 | 1.4 | 1.24 × 10−1 |
| nov-miR-2549-5p | 0.054 | 1.34 × 10−4 | 1.3 | 7.89 × 10−1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minutentag, I.W.; Seneda, A.L.; Barros-Filhos, M.C.; de Carvalho, M.; Souza, V.G.P.; Hasimoto, C.N.; Moraes, M.P.T.; Marchi, F.A.; Lam, W.L.; Reis, P.P.; et al. Discovery of Novel miRNAs in Colorectal Cancer: Potential Biological Roles and Clinical Utility. Non-Coding RNA 2023, 9, 65. https://doi.org/10.3390/ncrna9060065
Minutentag IW, Seneda AL, Barros-Filhos MC, de Carvalho M, Souza VGP, Hasimoto CN, Moraes MPT, Marchi FA, Lam WL, Reis PP, et al. Discovery of Novel miRNAs in Colorectal Cancer: Potential Biological Roles and Clinical Utility. Non-Coding RNA. 2023; 9(6):65. https://doi.org/10.3390/ncrna9060065
Chicago/Turabian StyleMinutentag, Iael Weissberg, Ana Laura Seneda, Mateus C. Barros-Filhos, Márcio de Carvalho, Vanessa G. P. Souza, Claudia N. Hasimoto, Marcelo P. T. Moraes, Fabio A. Marchi, Wan L. Lam, Patricia P. Reis, and et al. 2023. "Discovery of Novel miRNAs in Colorectal Cancer: Potential Biological Roles and Clinical Utility" Non-Coding RNA 9, no. 6: 65. https://doi.org/10.3390/ncrna9060065
APA StyleMinutentag, I. W., Seneda, A. L., Barros-Filhos, M. C., de Carvalho, M., Souza, V. G. P., Hasimoto, C. N., Moraes, M. P. T., Marchi, F. A., Lam, W. L., Reis, P. P., & Drigo, S. A. (2023). Discovery of Novel miRNAs in Colorectal Cancer: Potential Biological Roles and Clinical Utility. Non-Coding RNA, 9(6), 65. https://doi.org/10.3390/ncrna9060065