Novel Wine Yeast for Improved Utilisation of Proline during Fermentation
Abstract
:1. Introduction
2. Materials and Methods
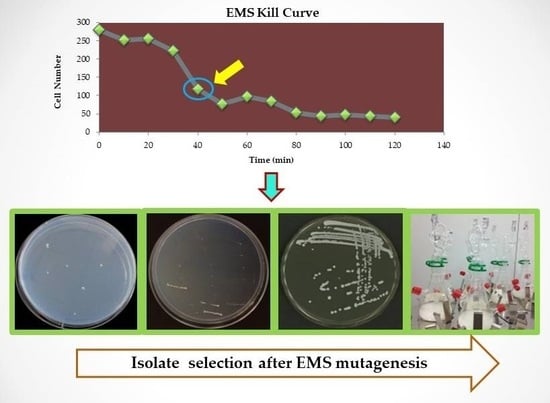
2.1. Yeast Strains and EMS Mutagenesis
2.2. Selection of Isolates
2.3. Influence of N Source on Growth of QA23 and EMS Isolate Q7
2.4. Yeast Performance during Fermentation
2.5. Extraction of Intracellular Proline from Yeast Cells
2.6. Freeze Tolerance Tests
2.7. Real-Time Quantitative PCR
2.7.1. RNA Extraction
2.7.2. Reverse Transcription and Real-Time PCR
2.8. Gene Sequencing
2.9. HPLC Analysis of Fermentation Metabolites
3. Results and Discussion
3.1. Comparison of Growth of QA23 and its EMS Isolate Q7
3.2. Fermentation Performance of QA23 and EMS Isolate Q7 in CDGJM.
3.3. Cell Morphology, Dry Weight and Intracellular Proline Content
3.4. Freeze Test
3.5. Analysis of Gene Expression
3.6. Gene Sequencing
3.7. Fermentation in Chenin Blanc Grape Juice
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ough, C.S.; Amerine, M.A. Methods for Analysis of Musts and Wines; Wiley: New York, NY, USA, 1988; ISBN 0471627577. [Google Scholar]
- Henschke, P.A.; Jiranek, V. Yeasts-metabolism of nitrogen compounds. In Wine Microbiology and Biotechnology; Harwood Academic Publishers GmbH: Amsterdam, The Netherlands, 1993; pp. 77–164. ISBN 978-0-41-527850-8. [Google Scholar]
- Alexandre, H.; Charpentier, C. Biochemical aspects of stuck and sluggish fermentation in grape must. J. Ind. Microbiol. Biotechnol. 1998, 20, 20–27. [Google Scholar] [CrossRef]
- Burin, V.M.; Gomes, T.M.; Caliari, V.; Rosier, J.P.; Bordignon Luiz, M.T. Establishment of influence the nitrogen content in musts and volatile profile of white wines associated to chemometric tools. Microchem. J. 2015, 122, 20–28. [Google Scholar] [CrossRef]
- Jiranek, V.; Langridge, P.; Henschke, P.A. Amino acid and ammonium utilization by Saccharomyces cerevisiae Wine yeasts from a chemically defined medium. Am. J. Enol. Vitic. 1995, 46, 75–83. [Google Scholar]
- Jiranek, V.; Langridge, P.; A Henschke, P. Regulation of hydrogen sulfide liberation in wine-producing Saccharomyces cerevisiae strains by assimilable nitrogen. Appl. Environ. Microbiol. 1995, 61, 461–467. [Google Scholar] [PubMed]
- Ingledew, W.M.; Kunkee, R.E. Factors influencing sluggish fermentations of grape juice. Am. J. Enol. Vitic. 1985, 36, 65–76. [Google Scholar]
- Monteiro, F.F.; Bisson, L.F. Biological assay of nitrogen content of grape juice and prediction of sluggish fermentations. Am. J. Enol. Vitic. 1991, 42, 47–57. [Google Scholar]
- Bisson, L.F. Stuck and sluggish fermentations. Am. J. Enol. Vitic. 1999, 50, 107–119. [Google Scholar]
- Gutiérrez, A.; Chiva, R.; Sancho, M.; Beltran, G.; Arroyo-López, F.N.; Guillamon, J.M. Nitrogen requirements of commercial wine yeast strains during fermentation of a synthetic grape must. Food Microbiol. 2012, 31, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, F.F.; Trousdale, E.K.; Bisson, L. Ethyl carbamate formation in wine: Use of radioactively labeled precursors to demonstrate the involvement of urea. Am. J. Enol. Vitic. 1989, 40, 1–8. [Google Scholar]
- Adams, C.; Vuuren, H. Effect of timing of diammonium phosphate addition to fermenting grape must on the production of ethyl carbamate in wine. Am. J. Enol. Vitic. 2010, 61, 125–129. [Google Scholar]
- Ugliano, M.; Henschke, P.A.; Herderich, M.; Pretorius, I.S. Nitrogen management is critical for wine flavour and style. Aust. N. Z. Wine Ind. J. 2007, 22, 24–30. [Google Scholar]
- Stines, A.P.; Grubb, J.; Gockowiak, H.; Henschke, P.A.; Høj, P.B.; van Heeswijck, R. Proline and arginine accumulation in developing berries of Vitis vinifera L. in Australian vineyards: Influence of vine cultivar, berry maturity and tissue type. Aust. J. Grape Wine Res. 2000, 6, 150–158. [Google Scholar] [CrossRef]
- Huang, Z.; Ough, C.S. Effect of vineyard locations, varieties, and rootstocks on the juice amino acid composition of severalcultivars. Am. J. Enol. Vitic. 1989, 40, 135–139. [Google Scholar]
- Brandriss, M.C.; Magasanik, B. Genetics and physiology of proline utilization in Saccharomyces cerevisiae: Enzyme induction by proline. J. Bacteriol. 1979, 140, 498–503. [Google Scholar] [PubMed]
- Wang, S.S.; Brandriss, M.C. Proline utilization in Saccharomyces cerevisiae: Sequence, regulation, and mitochondrial localization of the PUT1 gene product. Mol. Cell Biol. 1987, 7, 4431–4440. [Google Scholar] [CrossRef] [PubMed]
- Duteurtre, B.; Bourgeois, C.; Chollot, B. Study of the assimilation of proline by brewing yeast. J. Inst. Brew. 1971, 77, 28–35. [Google Scholar] [CrossRef]
- Takagi, H.; Sakai, K.; Morida, K.; Nakamori, S. Proline accumulation by mutation or disruption of the proline oxidase gene improves resistance to freezing and desiccation stresses in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 2000, 184, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H.; Taguchi, J.; Kaino, T. Proline accumulation protects Saccharomyces cerevisiae cells in stationary phase from ethanol stress by reducing reactive oxygen species levels. Yeast 2016, 33, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Ingledew, W.M.; Magnus, C.A.; Sosulski, F.W. Influence of oxygen on proline utilization during the wine fermentation. Am. J. Enol. Vitic. 1987, 38, 246–248. [Google Scholar]
- Sablayrolles, J.M.; Dubois, C.; Manginot, C.; Roustan, J.-L.; Barre, P. Effectiveness of combined ammoniacal nitrogen and oxygen additions for completion of sluggish and stuck wine fermentations. J. Ferment. Bioeng. 1996, 82, 377–381. [Google Scholar] [CrossRef]
- Gardner, J.M.; Poole, K.; Jiranek, V. Practical significance of relative assimilable nitrogen requirements of yeast: A preliminary study of fermentation performance and liberation of H2S. Aust. J. Grape Wine Res. 2002, 8, 175–179. [Google Scholar] [CrossRef]
- Omura, F.; Fujita, A.; Miyajima, K.; Fukui, N. Engineering of yeast PUT4 permease and its application to lager yeast for efficient proline assimilation. Biosci. Biotechnol. Biochem. 2005, 69, 1162–1171. [Google Scholar] [CrossRef] [PubMed]
- Poole, K.; Walker, M.E.; Warren, T.; Gardner, J.; McBryde, C.; de Barros Lopes, M.; Jiranek, V. Proline transport and stress tolerance of ammonia-insensitive mutants of the PUT4-encoded proline-specific permease in yeast. J. Gen. Appl. Microbiol. 2009, 55, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Liccioli, T. Improving Fructose Utilization in Wine Yeast Using Adaptive Evolution. Ph.D. Thesis, The University of Adelaide, Adelaide, Australia, 2010. Available online: http://hdl.handle.net/2440/67017 (accessed on 23 May 2013).
- Guthrie, C.; Fink, G.R. Guide to Yeast Genetics and Molecular Biology. Methods Enzymol. 1991, 194, 1–863. [Google Scholar]
- Poole, K. Enhancing Yeast Performance under Oenological Conditions by Enabling Proline Utilisation. Ph.D. Thesis, The University of Adelaide, Adelaide, Australia, 2002. Available online: http://hdl.handle.net/2440/58485 (accessed on 25 February 2011).
- Salmon, J.M.; Barre, P. Improvement of nitrogen assimilation and fermentation kinetics under enological conditions by derepression of alternative nitrogen-assimilatory pathways in an industrial Saccharomyces cerevisiae strain. Appl. Environ. Microbiol. 1998, 64, 3831–3837. [Google Scholar] [PubMed]
- Roon, R.J.; Even, H.L.; Dunlop, P.; Larimore, F.L. Methylamine and ammonia transport in Saccharomyces cerevisiae. J. Bacteriol. 1975, 122, 502–509. [Google Scholar] [PubMed]
- Long, D.; Wilkinson, K.L.; Poole, K.; Taylor, D.K.; Warren, T.; Astorga, A.M.; Jiranek, V. Rapid method for proline determination in grape juice and wine. J. Agric. Food Chem. 2012, 60, 4259–4264. [Google Scholar] [CrossRef] [PubMed]
- Boehringer Mannheim. D-glucose/D-fructose. In Methods of Biochemical Analysis and Food Analysis; Boehringer Mannheim: Mannheim, Germany, 1989; pp. 50–55. [Google Scholar]
- Ohsumi, Y.; Kitamoto, K.; Anraku, Y. Changes induced in the permeability barrier of the yeast plasma membrane by cupric ion. J. Bacteriol. 1988, 170, 2676–2682. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, K.; Takagi, H. Vacuolar functions are involved in stress-protective effect of intracellular proline in Saccharomyces cerevisiae. J. Biosci. Bioeng. 2005, 100, 538–544. [Google Scholar] [CrossRef] [PubMed]
- Kaino, T.; Takagi, H. Gene expression profiles and intracellular contents of stress protectants in Saccharomyces cerevisiae under ethanol and sorbitol stresses. Appl. Microbiol. Biotechnol. 2008, 79, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Beltran, G.; Novo, M.; Rozès, N.; Mas, A.; Guillamón, J.M. Nitrogen catabolite repression in Saccharomyces cerevisiae during wine fermentations. FEMS Yeast Res. 2004, 4, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Teste, M.-A.; Duquenne, M.; François, J.M.; Parrou, J.-L. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol. Biol. 2009, 10, 99. [Google Scholar] [CrossRef] [PubMed]
- Chomczynski, P.; Sacchi, N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: Twenty-something years on. Nat. Protoc. 2006, 1, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Ough, C.S.; Stashak, R.M. Further studies on proline concentration in grapes and wines. Am. J. Enol. Vitic. 1974, 25, 7–12. [Google Scholar]
- Lasko, P.F.; Brandriss, M.C. Proline transport in Saccharomyces cerevisiae. J. Bacteriol. 1981, 148, 241–247. [Google Scholar] [PubMed]
- Maggio, A.; Miyazaki, S.; Veronese, P.; Fujita, T.; Ibeas, J.I.; Damsz, B.; Narasimhan, M.L.; Hasegawa, P.M.; Joly, R.J.; Bressan, R.A. Does proline accumulation play an active role in stress-induced growth reduction? Plant J. 2002, 31, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Ciani, M.; Maccarelli, F.; Fatichenti, F. Growth and fermentation behaviour of Brettanomyces/Dekkera yeasts under different conditions of aerobiosis. World J. Microbiol. Biotechnol. 2003, 19, 419–422. [Google Scholar] [CrossRef]
- Aceituno, F.F.; Orellana, M.; Torres, J.; Mendoza, S.; Slater, A.W.; Melo, F.; Agosin, E. Oxygen response of the wine yeast Saccharomyces cerevisiae EC1118 grown under carbon-sufficient, nitrogen-limited enological conditions. Appl. Environ. Microbiol. 2012, 78, 8340–8352. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Nakamori, S.; Takagi, H. Effect of proline and arginine metabolism on freezing stress of Saccharomyces cerevisiae. J. Biosci. Bioeng. 2002, 94, 390–394. [Google Scholar] [CrossRef]
- Körber, C. Phenomena at the advancing ice-liquid interface: Solutes, particles and biological cells. Q. Rev. Biophys. 1988, 21, 229–298. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H.; Iwamoto, F.; Nakamori, S. Isolation of freeze-tolerant laboratory strains of Saccharomyces cerevisiae from proline-analogue-resistant mutants. Appl. Microbiol. Biotechnol. 1997, 47, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, A.H.; Brandriss, M.C. The Saccharomyces cerevisiae PUT3 activator protein associates with proline-specific upstream activation sequences. Mol. Cell Biol. 1989, 9, 4706–4712. [Google Scholar] [CrossRef] [PubMed]
- Axelrod, J.D.; Majors, J.; Brandriss, M.C. Proline-dependent binding of PUT3 transcriptional activator protein detected by footprinting in vivo. Mol. Cell Biol. 1991, 11, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Falvey, D.A.; Brandriss, M.C. Roles of URE2 and GLN3 in the proline utilization pathway in Saccharomyces cerevisiae. Mol. Cell Biol. 1995, 15, 2321–2330. [Google Scholar] [CrossRef] [PubMed]
- Daugherty, J.R.; Rai, R.; El Berry, H.M.; Cooper, T.G. Regulatory circuit for responses of nitrogen catabolic gene expression to the GLN3 and DAL80 proteins and nitrogen catabolite repression in Saccharomyces cerevisiae. J. Bacteriol. 1993, 175, 64–73. [Google Scholar] [CrossRef] [PubMed]







| Name | Primers (5′–3′) | Reference |
|---|---|---|
| PUT1-F | GATAAAACGGGCACTGACGA | [35] |
| PUT1-R | TAACGAGCATTTGGGATTGG | |
| PUT2-F | CCGATATGTTTGGCATATTGCA | [35] |
| PUT2-R | TGGACTTGCGGATGTGTTCA | |
| PUT3-F | GCGGTATTGAAGTCCTGTTGT | this study |
| PUT3-R | TGGATAGAGTGTCGCTTTGAGA | |
| PUT4-F | GAGCCGCACAAACTAAAACA | [35] |
| PUT4-R | CGTATGAAGCGTGGATGAAG | |
| GAP1-F | CTGTGGATGCTGCTGCTTCA | [36] |
| GAP1-R | CAACACTTGGCAAACCCTTGA | |
| URE2-F | TCCCGTATGGCTTGTAGGAGA | this study |
| URE2-R | CACGCAATGCCTTGATGACC | |
| ALG9-F | CACGGATAGTGGCTTTGGTGAACAATTAC | [37] |
| ALG9-R | TATGATTATCTGGCAGCAGGAAAGAACTTGGG | |
| TAF10-F | ATATTCCAGGATCAGGTCTTCCGTAGC | [37] |
| TAF10-R | GTAGTCTTCTCATTCTGTTGATGTTGTTGTTG |
| Strains | Starter | Aerobic | Self-Anaerobic | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cytosolic | Vacuolar | Cytosolic | Vacuolar | |||||||||||
| Cytosolic | Vacuolar | 24 h | mid | End | 24 h | mid | End | 24 h | mid | End | 24 h | mid | End | |
| QA23 | 17 ± 1 a | 132 ± 1 a | 116 ± 1 a | 10 ± 1 a | 1 ± 0 a | 136 ± 5 a | 94 ± 5 a | 16 ± 1 a | 122 ± 4 a | 13 ± 0 a | 5 ± 1 a | 47 ± 2 a | 64 ± 2 a | 4 ± 0 |
| Q7 | nd b | nd b | 81 ± 3 b | nd b | nd b | nd b | 1 ± 0 b | nd b | 90 ± 3 b | nd b | nd b | nd b | nd b | 4 ± 0 |
| Strain | −20 °C | −80 °C | ||
|---|---|---|---|---|
| Aerobic | Self-Anaerobic | Aerobic | Self-Anaerobic | |
| QA23 | 63 ± 6 a | 57 ± 5 a | 66 ± 6 a | 63 ± 5 a |
| Q7 | 20 ± 2 b | 29 ± 5 b | 29 ± 2 b | 37 ± 5 b |
| Gene | Strain | 55 (Promoter) | 260 (ORF) | Amino Acid | 580 (ORF) | Amino Acid |
|---|---|---|---|---|---|---|
| PUT1 | QA23 | G | G | Ser | C | Leu |
| Q7 | T | A | Asn | G | Val | |
| 45 (Promoter) | 465 (ORF) | Amino Acid | ||||
| PUT4 | QA23 | - | W | Thr/Ser | ||
| Q7 | - | T | Thr |
| Metabolites (g/L) | QA23 | Q7 |
|---|---|---|
| Malic acid | 3.1 ± 0.08 | 3.5 ± 0.22 |
| Citric acid | 3.5 ± 0.03 | 2.9 ± 0.36 |
| Lactic acid | 0.9 ± 0.05 | 0.9 ± 0.21 |
| Acetic acid | 0.2 ± 0.04 | 0.2 ± 0.07 |
| Succinic acid | 2.7 ± 0.04 | 3.2 ± 0.22 |
| Glycerol | 8.0 ± 0.08 | 7.7 ± 0.81 |
| Ethanol | 92.2 ± 0.41 | 85.7 ± 6.24 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Long, D.; Wilkinson, K.L.; Taylor, D.K.; Jiranek, V. Novel Wine Yeast for Improved Utilisation of Proline during Fermentation. Fermentation 2018, 4, 10. https://doi.org/10.3390/fermentation4010010
Long D, Wilkinson KL, Taylor DK, Jiranek V. Novel Wine Yeast for Improved Utilisation of Proline during Fermentation. Fermentation. 2018; 4(1):10. https://doi.org/10.3390/fermentation4010010
Chicago/Turabian StyleLong, Danfeng, Kerry L. Wilkinson, Dennis K. Taylor, and Vladimir Jiranek. 2018. "Novel Wine Yeast for Improved Utilisation of Proline during Fermentation" Fermentation 4, no. 1: 10. https://doi.org/10.3390/fermentation4010010
APA StyleLong, D., Wilkinson, K. L., Taylor, D. K., & Jiranek, V. (2018). Novel Wine Yeast for Improved Utilisation of Proline during Fermentation. Fermentation, 4(1), 10. https://doi.org/10.3390/fermentation4010010