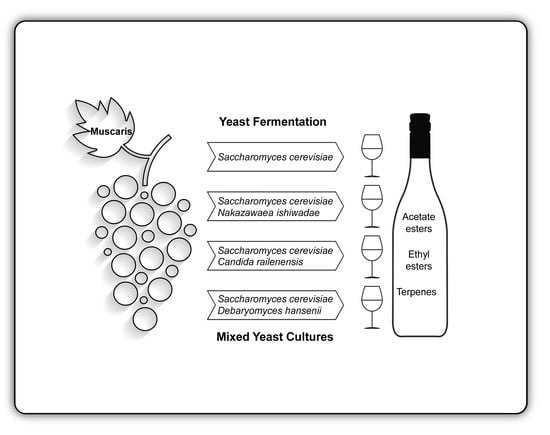
Assessing the Oenological Potential of Nakazawaea ishiwadae, Candida railenensis and Debaryomyces hansenii Strains in Mixed-Culture Grape Must Fermentation with Saccharomyces cerevisiae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Grape Must
2.2. Selection of Strains
2.3. Micro-Vinification
2.4. HPLC-Analysis
2.5. Volatile Compound Analysis
2.6. Statistical Analysis
3. Results and Discussion
3.1. Strain Identification
3.2. Fermentation
3.3. Volatile Composition
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Christ, E.; Kowalczyk, M.; Zuchowska, M.; Claus, H.; Löwenstein, R. An exemplary model study for overcoming stuck fermentation during spontaneous fermentation with the aid of a Saccharomyces triple hybrid. J. Agric. Sci. 2015, 7, 8. [Google Scholar] [CrossRef] [Green Version]
- Borneman, A.R.; Forgan, A.H.; Kolouchova, R.; Fraser, J.A.; Schmidt, S.A. Whole genome comparison reveals high levels of inbreeding and strain redundancy across the spectrum of commercial wine strains of Saccharomyces cerevisiae. G3 2016, 6, 957–971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Comitini, F.; Capece, A.; Ciani, M.; Romano, P. New insights on the use of wine yeasts. Curr. Opin. Food Sci. 2017, 13, 44–49. [Google Scholar] [CrossRef]
- Ciani, M.; Comitini, F.; Mannazzu, I.; Domizio, P. Controlled mixed culture fermentation: A new perspective on the use of non-Saccharomyces yeasts in winemaking. FEMS Yeast Res. 2010, 10, 123–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jolly, N.P.; Varela, C.; Pretorius, I.S. Not your ordinary yeast: Non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 2014, 14, 215–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Wyk, N.; Grossmann, M.; Wendland, J.; Von Wallbrunn, C.; Pretorius, I.S. The whiff of wine yeast innovation: Strategies for enhancing aroma production by yeast during wine fermentation. J. Agric. Food Chem. 2019, 67, 13496–13505. [Google Scholar] [CrossRef]
- Pretorius, I.S.; van der Westhuizen, T.J.; Augustyn, O.P.H. Yeast biodiversity in vineyards and wineries and its importance to the South African wine industry. S. Afr. J. Enol. Vitic. 1999, 20, 61–74. [Google Scholar] [CrossRef] [Green Version]
- Barata, A.; Malfeito-Ferreira, M.; Loureiro, V. The microbial ecology of wine grape berries. Int. J. Food Microbiol. 2012, 153, 243–259. [Google Scholar] [CrossRef]
- Shi, W.; Wang, J.; Chen, F.; Zhang, X. Effect of Issatchenkia terricola and Pichia kudriavzevii on wine flavor and quality through simultaneous and sequential co-fermentation with Saccharomyces cerevisiae. LWT Food Sci. Technol. 2019, 116, 108477. [Google Scholar] [CrossRef]
- Kim, D.H.; Hong, Y.A.; Park, H.D. Co-fermentation of grape must by Issatchenkia orientalis and Saccharomyces cerevisiae reduces the malic acid content in wine. Biotechnol. Lett. 2008, 30, 1633–1638. [Google Scholar] [CrossRef]
- Medina, K.; Boido, E.; Fariña, L.; Gioia, O.; Gomez, M.E.; Barquet, M.; Gaggero, C.; Dellacassa, E.; Carrau, F. Increased flavour diversity of Chardonnay wines by spontaneous fermentation and co-fermentation with Hanseniaspora vineae. Food Chem. 2013, 141, 2513–2521. [Google Scholar] [CrossRef] [PubMed]
- Rojas, V.; Gil, J.V.; Piñaga, F.; Manzanares, P. Studies on acetate ester production by non-Saccharomyces wine yeasts. Int. J. Food Microbiol. 2001, 70, 283–289. [Google Scholar] [CrossRef]
- Dukes, B.C.; Butzke, C.E. Rapid determination of primary amino acids in grape juice using an o-phthaldialdehyde/N-acetyl-L-cysteine spectrophotometric assay. Am. J. Enol. Vitic. 1998, 49, 125–134. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, A.; Gerbi, V.; Redoglia, M. A rapid HPLC method for separation and determination of major organic acids in grape musts and wines. Am. J. Enol. Vitic. 1987, 38, 151–155. Available online: https://www.ajevonline.org/content/ajev/38/2/151.full (accessed on 6 May 2020).
- Kanter, J.P.; Benito, S.; Brezina, S.; Beisert, B.; Fritsch, S.; Patz, C.D.; Rauhut, D. The impact of hybrid yeasts on the aroma profile of cool climate Riesling wines. Food Chem. X 2020, 5, 100072. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, J.; Belda, I.; Beisert, B.; Navascués, E.; Marquina, D.; Calderón, F.; Rauhut, D.; Santos, A.; Benito, S. Analytical impact of Metschnikowia pulcherrima in the volatile profile of Verdejo white wines. Appl. Microbiol. Biotechnol. 2018, 102, 8501–8509. [Google Scholar] [CrossRef] [Green Version]
- Varela, C.; Barker, A.; Tran, T.; Borneman, A.; Curtin, C. Sensory profile and volatile aroma composition of reduced alcohol Merlot wines fermented with Metschnikowia pulcherrima and Saccharomyces uvarum. Int. J. Food Microbiol. 2017, 252, 1–9. [Google Scholar] [CrossRef]
- Barbosa, C.; Lage, P.; Esteves, M.; Chambel, L.; Mendes-Faia, A.; Mendes-Ferreira, A. Molecular and phenotypic characterization of Metschnikowia pulcherrima strains from Douro Wine Region. Fermentation 2018, 4, 8. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Brysch-Herzberg, M.; Seidel, M. Yeast diversity on grapes in two German wine growing regions. Int. J. Food Microbiol. 2015, 214, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Cioch-Skoneczny, M.; Satora, P.; Skotniczny, M.; Skoneczny, S. Quantitative and qualitative composition of yeast microbiota in spontaneously fermented grape musts obtained from cool climate grape varieties ‘Rondo’ and ‘Regent’. FEMS Yeast Res. 2018, 18, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Drumonde-Neves, J.; Franco-Duarte, R.; Lima, T.; Schuller, D.; Pais, C. Association between Grape Yeast Communities and the Vineyard Ecosystems. PLoS ONE 2017, 12, e0169883. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, J.; Ortega, N.; Martín-Santamaría, M.; Acedo, A.; Marquina, D.; Pascual, O.; Rozès, N.; Zamora, F.; Santos, A.; Belda, I. Occurrence and enological properties of two new non-conventional yeasts (Nakazawaea ishiwadae and Lodderomyces elongisporus) in wine fermentations. Int. J. Food Microbiol. 2019, 305, 108255. [Google Scholar] [CrossRef] [PubMed]
- Belda, I.; Ruiz, J.; Esteban-Fernández, A.; Navascués, E.; Marquina, D.; Santos, A.; Moreno-Arribas, M.V. Microbial contribution to wine aroma and its intended use for wine quality improvement. Molecules 2017, 22, 189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Besancon, X.; Ratomahenina, R.; Galzy, P. Isolation and partial characterization of an esterase (EC 3.1.1.1) from a Debaryomyces hansenii strain. Neth. Milk Dairy J. 1995, 49, 97–110. [Google Scholar]
- Pedneault, K.; Provost, C. Fungus resistant grape varieties as a suitable alternative for organic wine production: Benefits, limits, and challenges. Sci. Hortic. (Amst.) 2016, 208, 57–77. [Google Scholar] [CrossRef]
- Yanai, T.; Sato, M. Isolation and properties of beta-glucosidase produced by Debaryomyces hansenii and its application in winemaking. Am. J. Enol. Vitic. 1999, 50, 231–235. [Google Scholar]



| Residual Sugars | Tartaric Acid | Malic Acid | Acetic Acid | Citric Acid | Lactic Acid | Shikimic Acid | Glycerol | Ethanol | |
|---|---|---|---|---|---|---|---|---|---|
| Vin13 (alone) | nd | 3.7 ± 0.0 | 3.1 ± 0.0 | 0.42 ± 0.0 | 0.22 ± 0.0 | 0.18 ± 0.0 | 0.008 ± 0.0 | 6.97 ± 0.1 | 12.57 ± 0.2 |
| Vin13 (+ C. railenensis) | nd | 4.0 ± 0.1 *** | 3.1 ± 0.0ns | 0.51 ± 0.0 ** | 0.21 ± 0.0ns | 0.17 ± 0.0ns | 0.008 ± 0.0ns | 6.90 ± 0.1ns | 12.73 ± 0.3ns |
| Vin13 (+ N. ishiwadae) | 2.4 ± 0.1 | 4.0 ± 0.1 ** | 3.0 ± 0.1ns | 0.45 ± 0.0ns | 0.22 ± 0.0ns | 0.17 ± 0.0ns | 0.002 ± 0.0 *** | 6.20 ± 0.1 *** | 12.70 ± 0.2ns |
| Vin13 (+ D. hansenii) | 2.1 ± 0.0 | 4.0 ± 0.1 *** | 3.0 ± 0.1 * | 0.52 ± 0.0 ** | 0.21 ± 0.0ns | 0.17 ± 0.0ns | 0.008 ± 0.0ns | 6.67 ± 0.2 * | 12.53 ± 0.2ns |
| Aroma Descriptor | Vin13 (alone) | Vin13 (+ C. railenensis) | Vin13 (+ N. ishiwadae) | Vin13 (+ D. hansenii) | |
|---|---|---|---|---|---|
| β-myrcene | Pepper, spicy | 6.47 ± 1.8 | 6.09 ± 1.9ns | 6.58 ± 2.1ns | 3.98 ± 3.5ns |
| limonene | Citrus, orange | 1.10 ± 0.1 | 1.02 ± 0.2ns | 1.06 ± 0.1ns | 1.12 ± 0.1ns |
| cis-rose oxide | Rose | 1.05 ± 0.0 | 2.11 ± 0.2 *** | 1.24 ± 0.0ns | 1.92 ± 0.1 *** |
| trans-rose oxide | Rose | 0.29 ± 0.0 | 0.56 ± 0.0 *** | 0.34 ± 0.0ns | 0.51 ± 0.0 *** |
| cis-linalool oxide | Floral | 83.20 ± 4.5 | 75.70 ± 3.4ns | 77.18 ± 2.6ns | 80 ± 8.5ns |
| trans-linalool oxide | Floral | 42.29 ± 1.8 | 37.74 ± 2.2ns | 41.86 ± 1.5ns | 39.63 ± 4.8ns |
| nerol oxide | Green, herbal | 12.82 ± 0.1 | 12.93 ± 0.7ns | 11.11 ± 0.2 * | 12.62 ± 0.8ns |
| vitispirane | Floral, fruity | 0.62 ± 0.0 | 0.60 ± 0.0 * | 0.61 ± 0.0ns | 0.66 ± 0.0 ** |
| linalool | Floral, rose | 75.00 ± 1.3 | 85.55 ± 4.8 * | 78.63 ± 0.4ns | 84.12 ± 4.6 * |
| hotrienol | Sweet, tropical | 148.40 ± 1.1 | 160.20 ± 9.1 * | 140.1 ± 4.2ns | 101.4 ± 7.5 ** |
| α-terpineol | Pine, terpene | 220.60 ± 5.5 | 238.60 ± 17.2ns | 217.3 ± 9.4ns | 153.6 ± 11.5 *** |
| citronellol | Floral, citrus | 2.34 ± 0.6 | 7.80 ± 0.3 *** | 5.49 ± 1.2 ** | 3.82 ± 0.4ns |
| β-damascenone | Fruity, floral | 0.79 ± 0.0 | 1.00 ± 0.1 ** | 1.04 ± 0.0 ** | 0.90 ± 0.1ns |
| Aroma Descriptor | Vin13 (alone) | Vin13 (+C. railenensis) | Vin13 (+N. ishiwadae) | Vin13 (+D. hansenii) | |
|---|---|---|---|---|---|
| Higher alcohols | |||||
| 2-Methyl-butanol (mg/L) | Roasted, winey | 68.67 ± 10.0 | 88.67 ± 2.9 * | 74.33 ± 11.2ns | 86.00 ± 4.4ns |
| Isobutanol (mg/L) | Ethereal, winey | 78.00 ± 16.6 | 64.67 ± 4.0ns | 78.33 ± 18.9ns | 65.33 ± 1.2ns |
| Isoamyl alcohol (mg/L) | Fusel, alcoholic | 335.3 ± 49.4 | 391.7 ± 15.0ns | 348.7 ± 55.6ns | 369.7 ± 13.6ns |
| Hexanol (mg/L) | Herbal, ethereal | 790.7 ± 74.1 | 882.0 ± 29.7ns | 616.0 ± 56.3 ** | 868.3 ± 7.4ns |
| 2-Phenyl-ethanol (mg/L) | Floral, rose | 26.00 ± 2.0 | 31.00 ± 1.7ns | 26.67 ± 3.1ns | 31.00 ± 3.5ns |
| Acetate esters | |||||
| Ethyl acetate (mg/L) | Ethereal, fruity | 99.00 ± 9.8 | 72.67 ± 5.5 * | 245.0 ± 14.0 *** | 76.33 ± 6.0 * |
| Isoamyl acetate (μg/L) | Banana, fruity | 4520 ± 410 | 1530 ± 170 *** | 5858 ± 192 *** | 1606 ± 148 *** |
| 2-Methylbutyl acetate (μg/L) | Overripe, fruity | 277.0 ± 24.0 | 153.0 ± 16.8 *** | 376.0 ± 10.6 *** | 170.3 ± 11.6 *** |
| Phenethyl acetate (μg/L) | Floral, rose | 214.7 ± 11.7 | 132.0 ± 4.4 *** | 311.0 ± 12.2 *** | 124.7 ± 4.7 *** |
| Hexyl acetate (μg/L) | Green, fruity | 71.7 ± 8.7 | 0.0 ± 0.0 *** | 99.6 ± 4.0 *** | 0.0 ± 0.0 *** |
| Medium-chain fatty acids | |||||
| Isovaleric acid (μg/L) | Cheesy, dairy | 1253 ± 6.4 | 1318 ± 14.6 *** | 1316 ± 15.0 *** | 1284 ± 3.2 *** |
| Hexanoic acid (mg/L) | Sour, fatty | 5.0 ± 0.0 | 4.0 ± 0.0ns | 5.0 ± 0.0ns | 5.0 ± 0.0ns |
| Octanoic acid (mg/L) | Fatty, waxy | 3.0 ± 0.0 | 3.0 ± 0.0ns | 3.0 ± 0.0ns | 2.3 ± 0.6ns |
| Decanoic acid (μg/L) | Sour, fatty | 864.7 ± 7.8 | 881.0 ± 10.2ns | 910.3 ± 6.7 *** | 871.7 ± 7.6ns |
| Ethyl esters | |||||
| Ethyl isobutyrate (μg/L) | Ethereal, fruity | 30.7 ± 1.5 | 36.7 ± 1.2 ** | 74.0 ± 2.6 *** | 41.3 ± 1.5 *** |
| Ethyl butyrate (μg/L) | Sweet, fruity | 625.3 ± 60.2 | 420.7 ± 42.1 *** | 737.7 ± 21.2 * | 415.0 ± 32.4 *** |
| Ethyl propionate (μg/L) | Sweet, fruity | 281.7 ± 25.3 | 354.3 ± 34.2 * | 690.0 ± 17.0 *** | 371.7 ± 37.0 * |
| Ethyl decanoate (μg/L) | Sweet, waxy | 334.0 ± 74.7 | 226.3 ± 54.1ns | 913.7 ± 43.1 *** | 370.0 ± 46.1ns |
| Ethyl octanoate (μg/L) | Fruity, winey | 953.3 ± 206.8 | 104.0 ± 93.0 *** | 1710 ± 76.0 *** | 727.3 ± 112.1ns |
| Ethyl hexanoate (μg/L) | Sweet, fruity | 870.7 ± 118.6 | 389.3 ± 60.7 *** | 1231 ± 40.3 ** | 538.7 ± 54.7 *** |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
van Wyk, N.; Pretorius, I.S.; von Wallbrunn, C. Assessing the Oenological Potential of Nakazawaea ishiwadae, Candida railenensis and Debaryomyces hansenii Strains in Mixed-Culture Grape Must Fermentation with Saccharomyces cerevisiae. Fermentation 2020, 6, 49. https://doi.org/10.3390/fermentation6020049
van Wyk N, Pretorius IS, von Wallbrunn C. Assessing the Oenological Potential of Nakazawaea ishiwadae, Candida railenensis and Debaryomyces hansenii Strains in Mixed-Culture Grape Must Fermentation with Saccharomyces cerevisiae. Fermentation. 2020; 6(2):49. https://doi.org/10.3390/fermentation6020049
Chicago/Turabian Stylevan Wyk, Niel, Isak S. Pretorius, and Christian von Wallbrunn. 2020. "Assessing the Oenological Potential of Nakazawaea ishiwadae, Candida railenensis and Debaryomyces hansenii Strains in Mixed-Culture Grape Must Fermentation with Saccharomyces cerevisiae" Fermentation 6, no. 2: 49. https://doi.org/10.3390/fermentation6020049
APA Stylevan Wyk, N., Pretorius, I. S., & von Wallbrunn, C. (2020). Assessing the Oenological Potential of Nakazawaea ishiwadae, Candida railenensis and Debaryomyces hansenii Strains in Mixed-Culture Grape Must Fermentation with Saccharomyces cerevisiae. Fermentation, 6(2), 49. https://doi.org/10.3390/fermentation6020049