Snapshot of Cyprus Raw Goat Milk Bacterial Diversity via 16S rDNA High-Throughput Sequencing; Impact of Cold Storage Conditions
Abstract
:1. Introduction
2. Materials and Methods
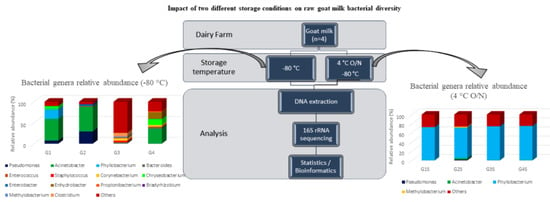
2.1. Sample Collection
2.2. Metagenomic DNA Extraction
2.3. Quantification of Total DNA
2.4. Barcoded Illumina MiSeq Amplicon Sequencing of Bacterial 16s rRNA Gene
2.5. Microbiome and Statistical Analysis
3. Results
3.1. Abundance and Diversity of Members of the Bacterial Microbiota
3.2. Taxonomic Composition of Bacterial Communities in Goat and Sheep Milk Samples
3.3. Relationships Between Milk Samples Bacterial Communities
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lad, S.S.; Aparnathi, K.D.; Mehta, B.; Velpula, S. Goat Milk in Human Nutrition and Health—A Review. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 1781–1792. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.W.; Juárez, M.; Ramos, M.; Haenlein, G.F.W. Physico-chemical characteristics of goat and sheep milk. Small Rumin. Res. 2007, 68, 88–113. [Google Scholar] [CrossRef] [Green Version]
- Kamilari, E.; Anagnostopoulos, D.A.; Papademas, P.; Kamilaris, A.; Tsaltas, D. Characterizing Halloumi cheese’s bacterial communities through metagenomic analysis. LWT 2020, 126, 109298. [Google Scholar] [CrossRef] [Green Version]
- Papademas, P.; Aspri, M.; Mariou, M.; Dowd, S.E.; Kazou, M.; Tsakalidou, E. Conventional and omics approaches shed light on Halitzia cheese, a long-forgotten white-brined cheese from Cyprus. Int. Dairy J. 2019, 98, 72–83. [Google Scholar] [CrossRef]
- Park, Y.W. Goat Milk: Goat Milk-Chemistry and Nutrition. In Handbook of Milk of Non-Bovine Mammals; Park, Y.W., Haenlein, G.F.W., Eds.; Blackwell Publishing Professional: Oxford, UK; Ames, IA, USA, 2006; pp. 34–58. [Google Scholar]
- Tagliazucchi, D.; Martini, S.; Shamsia, S.; Helal, A.; Conte, A. Biological activities and peptidomic profile of in vitro-digested cow, camel, goat and sheep milk. Int. Dairy J. 2018, 81, 19–27. [Google Scholar] [CrossRef] [Green Version]
- Capriotti, A.L.; Cavaliere, C.; Piovesana, S.; Samperi, R.; Laganà, A. Recent trends in the analysis of bioactive peptides in milk and dairy products. Anal. Bioanal. Chem. 2016, 408, 2677–2685. [Google Scholar] [CrossRef]
- Marcone, S.; Belton, O.; Fitzgerald, D.J. Milk-derived bioactive peptides and their health promoting effects: A potential role in atherosclerosis. Br. J. Clin. Pharmacol. 2017, 83, 152–162. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, S.D.; Beverly, R.L.; Qu, Y.; Dallas, D.C. Milk bioactive peptide database: A comprehensive database of milk protein-derived bioactive peptides and novel visualization. Food Chem. 2017, 232, 673–682. [Google Scholar] [CrossRef]
- Griffiths, M.I.W.; Laing, R.R.; Roy, D.; Mafu, A.A. Psychrotrophs in dairy products: Their effects and their control. Crit. Rev. Food Sci. Nutr. 1994, 34, 1–30. [Google Scholar] [CrossRef]
- Hantsis-Zacharov, E.; Halpern, M. Culturable psychrotrophic bacterial communities in raw milk and their proteolytic and lipolytic traits. Appl. Environ. Microbiol. 2007, 73, 7162–7168. [Google Scholar] [CrossRef] [Green Version]
- Alessandria, V.; Ferrocino, I.; De Filippis, F.; Fontana, M.; Rantsiou, K.; Ercolini, D.; Cocolin, L. Microbiota of an Italian Grana-like cheese during manufacture and ripening, unraveled by 16S rRNA-based approaches. Appl. Environ. Microbiol. 2016, 82, 3988–3995. [Google Scholar] [CrossRef] [Green Version]
- Quigley, L.; O’Sullivan, O.; Stanton, C.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. The complex microbiota of raw milk. FEMS Microbiol. Rev. 2013, 37, 664–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, F.; Liu, S.; Zhou, X.; Wang, P.; He, R.; Zhou, Z.; Zou, C. High-throughput sequencing analysis of bacterial diversity in raw and pasteurized goat milk. BioRxiv 2019, 751149. [Google Scholar] [CrossRef]
- McInnis, E.A.; Kalanetra, K.M.; Mills, D.A.; Maga, E.A. Analysis of raw goat milk microbiota: Impact of stage of lactation and lysozyme on microbial diversity. Food Microbiol. 2015, 46, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, Z.; Lei, F.; Wang, B.; Jiang, S.; Peng, Q.; Zhang, J.; Shao, Y. Bacterial diversity in goat milk from the Guanzhong area of China. J. Dairy Sci. 2017, 100, 7812–7824. [Google Scholar] [CrossRef] [Green Version]
- Di Bella, J.M.; Bao, Y.; Gloor, G.B.; Burton, J.P.; Reid, G. High throughput sequencing methods and analysis for microbiome research. J. Microbiol. Methods 2013, 95, 401–414. [Google Scholar] [CrossRef]
- De Filippis, F.; Parente, E.; Ercolini, D. Metagenomics insights into food fermentations. Microb. Biotechnol. 2017, 10, 91–102. [Google Scholar] [CrossRef]
- Loman, N.J.; Constantinidou, C.; Chan, J.Z.M.; Halachev, M.; Sergeant, M.; Penn, C.W.; Robinson, E.R.; Pallen, M.J. High-throughput bacterial genome sequencing: An embarrassment of choice, a world of opportunity. Nat. Rev. Microbiol. 2012, 10, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M. Usadel, BTrimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics 2014, btu170. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macklin, M.T.; Mann, H.B. Fallacies inherent in the proband method of analysis of human pedigrees. Am. J. Dis. Child. 1947, 74, 456–467. [Google Scholar] [CrossRef]
- Kamilari, E.; Tomazou, M.; Antoniades, A.; Tsaltas, D. High throughput sequencing technologies as a new toolbox for deep analysis, characterization and potentially authentication of protection designation of origin cheeses? Int. J. Food Sci. 2019, 2019, 5837301. [Google Scholar] [CrossRef] [Green Version]
- Oikonomou, G.; Addis, M.F.; Chassard, C.; Nader-Macias, M.E.F.; Grant, I.; Delbès, C.; Bogni, C.I.; Le Loir, Y.; Even, S. Milk Microbiota: What Are We Exactly Talking About? Front. Microbiol. 2020, 11, 60. [Google Scholar] [CrossRef] [Green Version]
- Quigley, L.; McCarthy, R.; O’Sullivan, O.; Beresford, T.P.; Fitzgerald, G.F.; Ross, R.P.; Stanton, C.; Cotter, P.D. The microbial content of raw and pasteurized cow milk as determined by molecular approaches. J. Dairy Sci. 2013, 96, 4928–4937. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Zhang, Y.; Liu, H.; Zhao, S.; Wang, J.; Zheng, N. Characterization of Pseudomonas spp. and associated proteolytic properties in raw milk stored at low temperatures. Front. Microbiol. 2017, 8, 2158. [Google Scholar] [CrossRef] [Green Version]
- Gurung, M.; Nam, H.M.; Tamang, M.D.; Chae, M.H.; Jang, G.C.; Jung, S.C.; Lim, S.K. Prevalence and antimicrobial susceptibility of Acinetobacter from raw bulk tank milk in Korea. J. Dairy Sci. 2013, 96, 1997–2002. [Google Scholar] [CrossRef]
- Esquivel-Elizondo, S.; Ilhan, Z.E.; Garcia-Pena, E.I.; Krajmalnik-Brown, R. Insights into butyrate production in a controlled fermentation system via gene predictions. mSystems 2017, 2, 13. [Google Scholar] [CrossRef] [Green Version]
- Den Besten, G.; Van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russo, P.; Fiocco, D.; Albenzio, M.; Spano, G.; Capozzi, V. Microbial Populations of Fresh and Cold Stored Donkey Milk by High-Throughput Sequencing Provide Indication for A Correct Management of This High-Value Product. Appl. Sci. 2020, 10, 2314. [Google Scholar] [CrossRef] [Green Version]
- Raats, D.; Offek, M.; Minz, D.; Halpern, M. Molecular analysis of bacterial communities in raw cow milk and the impact of refrigeration on its structure and dynamics. Food Microbiol. 2011, 28, 465–471. [Google Scholar] [CrossRef]
- Mantelin, S.; Fischer-Le Saux, M.; Zakhia, F.; Béna, G.; Bonneau, S.; Jeder, H.; de Lajudie, P.; Cleyet-Marel, J.C. Emended description of the genus Phyllobacterium and description of four novel species associated with plant roots: Phyllobacterium bourgognense sp. nov., Phyllobacterium ifriqiyense sp. nov., Phyllobacterium leguminum sp. nov. and Phyllobacterium brassic. Int. J. Syst. Evol. Microbiol. 2006, 56, 827–839. [Google Scholar] [CrossRef] [Green Version]
- Lambert, B.; Joos, H.; Dierickx, S.; Vantomme, R.; Swings, J.; Kersters, K.; Van Montagu, M. Identification and plant interaction of a Phyllobacterium sp., a predominant rhizobacterium of young sugar beet plants. Appl. Environ. Microbiol. 1990, 56, 1093–1102. [Google Scholar] [CrossRef] [Green Version]
- Mergaert, J.; Cnockaert, M.C.; Swings, J. Phyllobacterium myrsinacearum (subjective synonym Phyllobacterium rubiacearum) emend. Int. J. Syst. Evol. Microbiol. 2002, 52, 1821–1823. [Google Scholar] [CrossRef]
- Bertrand, H.; Nalin, R.; Bally, R.; Cleyet-Marel, J.C. Isolation and identification of the most efficient plant growth-promoting bacteria associated with canola (Brassica napus). Biol. Fertil. Soils 2001, 33, 152–156. [Google Scholar] [CrossRef]
- Larcher, M.; Muller, B.; Mantelin, S.; Rapior, S.; Cleyet-Marel, J.C. Early modifications of Brassica napus root system architecture induced by a plant growth-promoting Phyllobacterium strain. New Phytol. 2003, 160, 119–125. [Google Scholar] [CrossRef]
- Mantelin, S.; Desbrosses, G.; Larcher, M.; Tranbarger, T.J.; Cleyet-Marel, J.C.; Touraine, B. Nitrate-dependent control of root architecture and N nutrition are altered by a plant growth-promoting Phyllobacterium sp. Planta 2006, 223, 591–603. [Google Scholar] [CrossRef]
- Teh, G.K.; Flint, S.; Brooks, J.; Knight, G. Raw milk quality influenced by biofilms and the effect of biofilm growth on dairy product quality. In Biofilms in the Dairy Industry; Teh, K.H., Flint, S., Brooks, J., Knight, G., Eds.; Wiley: Chichester, UK, 2015; pp. 65–98. [Google Scholar] [CrossRef]
- Vithanage, N.R.; Yeager, T.R.; Jadhav, S.R.; Palombo, E.A.; Datta, N. Comparison of identification systems for psychrotrophic bacteria isolated from raw bovine milk. Int. J. Food Microbiol. 2014, 189, 26–38. [Google Scholar] [CrossRef]
- Doyle, C.J.; Gleeson, D.; O’Toole, P.W.; Cotter, P.D. High-throughput metataxonomic characterization of the raw milk microbiota identifies changes reflecting lactation stage and storage conditions. Int. J. Food Microbiol. 2017, 255, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Porcellato, D.; Aspholm, M.; Skeie, S.B.; Monshaugen, M.; Brendehaug, J.; Mellegård, H. Microbial diversity of consumption milk during processing and storage. Int. J. Food Microbiol. 2018, 266, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Cancino-Padilla, N.; Fellenberg, M.A.; Franco, W.; Ibáñez, R.A.; Vargas-Bello-Pérez, E. Foodborne bacteria in dairy products: Detection by molecular techniques. Cienc. Investig. Agrar. 2017, 44, 215–229. [Google Scholar] [CrossRef] [Green Version]
- De Filippis, F.; La Storia, A.; Stellato, G.; Gatti, M.; Ercolini, D. A selected core microbiome drives the early stages of three popular Italian cheese manufactures. PLoS ONE 2014, 9, e89680. [Google Scholar] [CrossRef] [PubMed]
- Yeluri Jonnala, B.R.; McSweeney, P.L.H.; Sheehan, J.J.; Cotter, P.D. Sequencing of the Cheese Microbiome and Its Relevance to Industry. Front. Microbiol. 2018, 9, 1020. [Google Scholar] [CrossRef] [Green Version]



| Sample Name | Area of Production | Type of Milk | Breed | Number of Animals | Feeding System |
|---|---|---|---|---|---|
| G1 | Pareklisia/Limassol | Goat | Macheras | 12 | Semi-extensive farming |
| G2 | Kritou Marotou/Paphos | Goat | Macheras | 30 | Semi-extensive farming |
| G3 | Kampia/Nicosia | Goat | Alpine | 3 | Semi-extensive farming |
| G4 | Anogyra/Limassol | Goat | Damascus | 26 | Semi-extensive farming |
| Sample-ID | Filtered Reads | Raw Reads | Shannon | Simpson | Chao1 | Observed OTUs |
|---|---|---|---|---|---|---|
| G1 | 15,744 | 29,480 | 3.454846 | 0.892831 | 23 | 20 |
| G1S | 30,359 | 59,132 | 2.944104 | 0.829086 | 62 | 42 |
| G2 | 8047 | 14,181 | 4.108257 | 0.922543 | 35 | 35 |
| G2S | 8293 | 16,347 | 3.117575 | 0.848981 | 33 | 33 |
| G3 | 14,755 | 38,065 | 8.598082 | 0.995163 | 1111 | 824 |
| G3S | 24,740 | 54,450 | 2.756315 | 0.801392 | 75 | 44 |
| G4 | 15,988 | 37,402 | 5.707971 | 0.956729 | 219 | 196 |
| G4S | 14,066 | 28,321 | 2.811905 | 0.811581 | 75 | 56 |
| Type of Milk | Country | Relative Abundance | Reference | ||
|---|---|---|---|---|---|
| ≥25% | 10–24% | 1–9% | |||
| Goat Macheras (n = 12) | Limassol/ Cyprus | Acinetobacter | Pseudomonas, Phyllobacterium | Chryseobacterium | Present study |
| Goat Macheras (n = 30) | Paphos/ Cyprus | Acinetobacter, Pseudomonas | - | - | |
| Goat Alpine (n = 3) | Nicosia/ Cyprus | - | - | Bacteroides, Staphylococcus, Corynebacterium, Methylobacterium, Clostridium | |
| Goat Damascus (n = 26) | Limassol/Cyprus | Acinetobacter | Chryseobacterium, Enhydrobacter | Bacteroides, Corynebacterium | |
| Goat Macheras (n = 10) | Paphos/Cyprus | Lactococcus, Leuconostoc | Pseudomonas | Carnobacterium Pahnella | [4] |
| Goat Guanzhong (n = 200) | Guangxi Zhuang/China | Kluyvera | Geobacillus, Thermus, Pseudomonas, Acinetobacter, Shigella, Aquabacterium, Burkholderia, Streptococcus | [14] | |
| Goat Alpine, Toggenburg, Saanen, LaMancha (n = 8) | United States | Pseudomonas | Rhodococcus | Micrococcus, Stenotrophomonas, Phyllobacterium, Streptococcus, Agrobacterium | [15] |
| Goat Saanen (n = 3) | China | Enterobacter | - | Pseudomonas, Acinetobacter Staphylococcus, Massilia, Bacillus, Streptococcus, Bacteroides | [16] |
| Goat Guanzhong (n = 3) | - | Pseudomonas, Acinetobacter, Enterobacter | Staphylococcus, Stenotrophomonas, Massilia, Bacillus, Streptococcus | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamilari, E.; Anagnostopoulos, D.A.; Papademas, P.; Efthymiou, M.; Tretiak, S.; Tsaltas, D. Snapshot of Cyprus Raw Goat Milk Bacterial Diversity via 16S rDNA High-Throughput Sequencing; Impact of Cold Storage Conditions. Fermentation 2020, 6, 100. https://doi.org/10.3390/fermentation6040100
Kamilari E, Anagnostopoulos DA, Papademas P, Efthymiou M, Tretiak S, Tsaltas D. Snapshot of Cyprus Raw Goat Milk Bacterial Diversity via 16S rDNA High-Throughput Sequencing; Impact of Cold Storage Conditions. Fermentation. 2020; 6(4):100. https://doi.org/10.3390/fermentation6040100
Chicago/Turabian StyleKamilari, Eleni, Dimitrios A. Anagnostopoulos, Photis Papademas, Marina Efthymiou, Svitlana Tretiak, and Dimitrios Tsaltas. 2020. "Snapshot of Cyprus Raw Goat Milk Bacterial Diversity via 16S rDNA High-Throughput Sequencing; Impact of Cold Storage Conditions" Fermentation 6, no. 4: 100. https://doi.org/10.3390/fermentation6040100
APA StyleKamilari, E., Anagnostopoulos, D. A., Papademas, P., Efthymiou, M., Tretiak, S., & Tsaltas, D. (2020). Snapshot of Cyprus Raw Goat Milk Bacterial Diversity via 16S rDNA High-Throughput Sequencing; Impact of Cold Storage Conditions. Fermentation, 6(4), 100. https://doi.org/10.3390/fermentation6040100