A Human and Animal Based Study Reveals That a Traditionally Fermented Rice Beverage Alters Gut Microbiota and Fecal Metabolites for Better Gut Health
Abstract
:1. Introduction
2. Materials and Methods
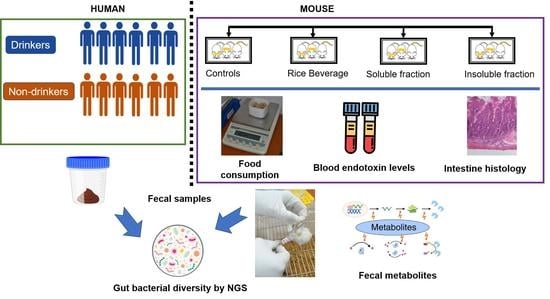
2.1. Recruitment of Human Subjects and Collection of Samples
2.2. Rice Fermentation
2.3. Mouse Experiment
2.4. Isolation of Fecal Metagenomic DNA and Next-Generation Sequencing (NGS) Analyses
2.5. Nontargeted Fecal Metabolite Profiling of Mice Feces via Gas Chromatography–Mass Spectrometry (GC–MS)
2.6. Assay of Plasma Endotoxin
2.7. Histology of Ileum
2.8. Statistical Analyses
3. Results
3.1. Dietary and Anthropometric Features of the Study Subjects
3.2. Gut Bacterial Diversity of Humans and Mice via NGS
3.3. Fecal Metabolites after 30 Days of Rice Beverage Treatment
3.4. Plasma Endotoxin and Histological Study of Ileum
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ASV | amplicon sequence variant |
| GC-MS | Gas chromatography–mass spectrometry |
| NGS | Next-generation sequencing |
| PLS–DA | Partial least squares discriminant analysis |
| SCFA | Short-chain fatty acids |
| DADA2 | Divisive amplicon denoising algorithm 2 |
| PCoA | Principal coordinate analysis |
References
- Guarner, F.; Malagelada, J.R. Gut flora in health and disease. Lancet 2003, 361, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Foster, K.R.; Schluter, J.; Coyte, K.Z.; Rakoff-Nahoum, S. The evolution of the host microbiome as an ecosystem on a leash. Nature 2017, 548, 43–51. [Google Scholar] [CrossRef] [Green Version]
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef] [Green Version]
- Dehingia, M.; Talukdar, N.C.; Talukdar, R.; Reddy, N.; Mande, S.S.; Deka, M.; Khan, M.R. Gut bacterial diversity of the tribes of India and comparison with the worldwide data. Sci. Rep. 2015, 5, 18563. [Google Scholar] [CrossRef] [Green Version]
- Ramos, S.; Martín, M.Á. Impact of diet on gut microbiota. Curr. Opin. Food Sci. 2021, 37, 83–90. [Google Scholar] [CrossRef]
- Wastyk, H.C.; Fragiadakis, G.K.; Perelman, D.; Dahan, D.; Merrill, B.D.; Feiqiao, B.Y.; Topf, M.; Gonzalez, C.G.; Van Treuren, W.; Han, S.; et al. Gut-microbiota-targeted diets modulate human immune status. Cell 2021, 184, 4137–4153. [Google Scholar] [CrossRef] [PubMed]
- Marco, M.L.; Heeney, D.; Binda, S.; Cifelli, C.J.; Cotter, P.D.; Foligné, B.; Gänzle, M.; Kort, R.; Pasin, G.; Pihlanto, A.; et al. Health benefits of fermented foods: Microbiota and beyond. Curr. Opin. Biotechnol. 2017, 44, 94–102. [Google Scholar] [CrossRef]
- Derrien, M.; van Hylckama Vlieg, J.E. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol. 2015, 23, 354–366. [Google Scholar] [CrossRef] [Green Version]
- Tamang, J.P. “Ethno-microbiology” of ethnic Indian fermented foods and alcoholic beverages. J. Appl. Microbiol. 2021, 133, 145–161. [Google Scholar] [CrossRef]
- Das, A.J.; Khawas, P.; Miyaji, T.; Deka, S.C. HPLC and GC-MS analyses of organic acids, carbohydrates, amino acids and volatile aromatic compounds in some varieties of rice beer from northeast India. J. Inst. Brew. 2014, 120, 244–252. [Google Scholar] [CrossRef]
- Roy, A.; Khanra, K.; Mishra, A.; Bhattacharyya, N. General analysis and antioxidant study of traditional fermented drink Handia, its concentrate and volatiles. Adv. Life Sci. Its Appl. 2012, 1, 54–57. [Google Scholar]
- Ghosh, K.; Ray, M.; Adak, A.; Dey, P.; Halder, S.K.; Das, A.; Jana, A.; Parua, S.; Mohapatra, P.K.D.; Pati, B.R.; et al. Microbial, saccharifying and antioxidant properties of an Indian rice based fermented beverage. Food Chem. 2015, 168, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Deka, S.; Miyaji, T. Methodology of rice beer preparation and various plant materials used in starter culture preparation by some tribal communities of North-East India: A survey. Int. Food Res. J. 2012, 19, 101. [Google Scholar]
- Bhaskar, B.; Adak, A.; Khan, M.R. Fractions of traditionally brewed rice beverage relieve anxiety and improve spatial memory in mice. J. Ethn. Foods 2021, 8, 1–9. [Google Scholar] [CrossRef]
- Das, S.; Deb, D.; Adak, A.; Khan, M.R. Exploring the microbiota and metabolites of traditional rice beer varieties of Assam and their functionalities. 3Biotech 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Fuloria, S.; Mehta, J.; Talukdar, M.P.; Sekar, M.; Gan, S.H.; Subramaniyan, V.; Rani, N.N.I.M.; Begum, M.Y.; Chidambaram, K.; Nordin, R.; et al. Synbiotic Effects of Fermented Rice on Human Health and Wellness: A Natural Beverage That Boosts Immunity. Front. Microbiol. 2022, 13, 1–20. [Google Scholar] [CrossRef]
- Khan, M.R.; Bhaskar, B.; Adak, A.; Talukdar, N.C. Rice based beverage with high alcohol content and method therefore. Pat. Off. J. India 2017, 11, 6850. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Wickham, H.; Chang, W.; Wickham, M.H. Package ‘ggplot2’. In Create Elegant Data Visualisations Using the Grammar of Graphics. Version; 2016; Volume 2, pp. 1–189. Available online: https://cran.microsoft.com/snapshot/2015-01-06/web/packages/ggplot2/ggplot2.pdf (accessed on 1 January 2023).
- Kassambara, A.; Kassambara, M.A. Package ‘ggpubr’. R Package Version 0.1. 2020. Available online: https://cran.microsoft.com/snapshot/2017-02-26/web/packages/ggpubr/ggpubr.pdf (accessed on 1 January 2023).
- Weir, T.L.; Manter, D.K.; Sheflin, A.M.; Barnett, B.A.; Heuberger, A.L.; Ryan, E.P. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS ONE 2013, 8, e70803. [Google Scholar] [CrossRef] [Green Version]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef]
- Zhou, Q.Q.; Yang, D.Z.; Luo, Y.J.; Li, S.Z.; Liu, F.Y.; Wang, G.S. Over-starvation aggravates intestinal injury and promotes bacterial and endotoxin translocation under high-altitude hypoxic environment. World J. Gastroenterol. WJG 2011, 17, 1584. [Google Scholar] [CrossRef] [PubMed]
- Precup, G.; Vodnar, D.C. Gut Prevotella as a possible biomarker of diet and its eubiotic versus dysbiotic roles: A comprehensive literature review. Br. J. Nutr. 2019, 122, 131–140. [Google Scholar] [CrossRef] [PubMed]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [Green Version]
- Amato, K.R.; Yeoman, C.J.; Cerda, G.; A Schmitt, C.; Cramer, J.D.; Miller, M.E.B.; Gomez, A.; R Turner, T.; Wilson, B.A.; Stumpf, R.M.; et al. Variable responses of human and non-human primate gut microbiomes to a Western diet. Microbiome 2015, 3, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engen, P.A.; Green, S.J.; Voigt, R.M.; Forsyth, C.B.; Keshavarzian, A. The gastrointestinal microbiome: Alcohol effects on the composition of intestinal microbiota. Alcohol Res. Curr. Rev. 2015, 37, 223. [Google Scholar]
- Dolara, P.; Luceri, C.; De Filippo, C.; Femia, A.P.; Giovannelli, L.; Caderni, G.; Cecchini, C.; Silvi, S.; Orpianesi, C.; Cresci, A. Red wine polyphenols influence carcinogenesis, intestinal microflora, oxidative damage and gene expression profiles of colonic mucosa in F344 rats. Mutat. Res./Fundam. Mol. Mech. Mutagen. 2005, 591, 237–246. [Google Scholar] [CrossRef]
- Kuprys, P.V.; Cannon, A.R.; Shieh, J.; Iftekhar, N.; Park, S.K.; Eberhardt, J.M.; Ding, X.; Choudhry, M.A. Alcohol decreases intestinal ratio of Lactobacillus to Enterobacteriaceae and induces hepatic immune tolerance in a murine model of DSS-colitis. Gut Microbes 2020, 12, 1838236. [Google Scholar] [CrossRef]
- Liang, C.; Zhou, X.H.; Jiao, Y.H.; Guo, M.J.; Meng, L.; Gong, P.M.; Lyu, L.Z.; Niu, H.Y.; Wu, Y.F.; Chen, S.W.; et al. Ligilactobacillus Salivarius LCK11 Prevents Obesity by Promoting PYY Secretion to Inhibit Appetite and Regulating Gut Microbiota in C57BL/6J Mice. Mol. Nutr. Food Res. 2021, 65, 2100136. [Google Scholar] [CrossRef]
- Yao, M.; Lu, Y.; Zhang, T.; Xie, J.; Han, S.; Zhang, S.; Fei, Y.; Ling, Z.; Wu, J.; Hu, Y.; et al. Improved functionality of Ligilactobacillus salivarius Li01 in alleviating colonic inflammation by layer-by-layer microencapsulation. NPJ Biofilms Microbiomes 2021, 7, 1–10. [Google Scholar] [CrossRef]
- Fei, Y.; Zhang, S.; Han, S.; Qiu, B.; Lu, Y.; Huang, W.; Li, F.; Chen, D.; Berglund, B.; Xiao, H.; et al. The Role of Dihydroresveratrol in Enhancing the Synergistic Effect of Ligilactobacillus salivarius Li01 and Resveratrol in Ameliorating Colitis in Mice. Research 2022, 2022, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Albarracin, L.; Indo, Y.; Arce, L.; Masumizu, Y.; Tomokiyo, M.; Islam, M.A.; Garcia-Castillo, V.; Ikeda-Ohtsubo, W.; Nochi, T.; et al. Selection of Immunobiotic Ligilactobacillus salivarius strains from the intestinal tract of wakame-fed pigs: Functional and genomic studies. Microorganisms 2020, 8, 1659. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Shen, Y.; Chen, M.; Zhang, Z.; Xiao, S.; Liu, C.; Wan, Y.; Yang, L.; Jiang, S.; Shang, E.; et al. Lizhong decoction ameliorates ulcerative colitis in mice via modulating gut microbiota and its metabolites. Appl. Microbiol. Biotechnol. 2020, 104, 5999–6012. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liu, W.; Feng, Y.; Hou, H.; Zhang, Z.; Yu, Q.; Zhou, Y.; Luo, Q.; Luo, Y.; Ouyang, H.; et al. Radix Puerariae thomsonii polysaccharide (RPP) improves inflammation and lipid peroxidation in alcohol and high-fat diet mice by regulating gut microbiota. Int. J. Biol. Macromol. 2022, 209, 858–870. [Google Scholar] [CrossRef]
- Song, X.; Zhong, L.; Lyu, N.; Liu, F.; Li, B.; Hao, Y.; Xue, Y.; Li, J.; Feng, Y.; Ma, Y.; et al. Inulin can alleviate metabolism disorders in ob/ob mice by partially restoring leptin-related pathways mediated by gut microbiota. Genom. Proteom. Bioinform. 2019, 17, 64–75. [Google Scholar] [CrossRef]
- Surono, I.S.; Wardana, A.A.; Waspodo, P.; Saksono, B.; Verhoeven, J.; Venema, K. Effect of functional food ingredients on gut microbiota in a rodent diabetes model. Nutr. Metab. 2020, 17, 1–9. [Google Scholar] [CrossRef]
- Wu, M.R.; Chou, T.S.; Huang, C.Y.; Hsiao, J.K. A Potential Probiotic-Lachnospiraceae NK4A136 Group: Evidence from the Restoration of the Dietary Pattern from a High-Fat Diet; Research Square: Tsukuba, Japan, 2020. [Google Scholar]
- Galié, S.; García-Gavilán, J.; Camacho-Barcía, L.; Atzeni, A.; Muralidharan, J.; Papandreou, C.; Arcelin, P.; Palau-Galindo, A.; Garcia, D.; Basora, J.; et al. Effects of the mediterranean diet or nut consumption on gut microbiota composition and fecal metabolites and their relationship with cardiometabolic risk factors. Mol. Nutr. Food Res. 2021, 65, 2000982. [Google Scholar] [CrossRef]
- Zhang, X.; Yasuda, K.; Gilmore, R.A.; Westmoreland, S.V.; Platt, D.M.; Miller, G.M.; Vallender, E.J. Alcohol-induced changes in the gut microbiome and metabolome of rhesus macaques. Psychopharmacology 2019, 236, 1531–1544. [Google Scholar] [CrossRef]
- Li, S.; Wang, Z.; Yang, Y.; Yang, S.; Yao, C.; Liu, K.; Cui, S.; Zou, Q.; Sun, H.; Guo, G. Lachnospiraceae shift in the microbial community of mice fecal sample effects on water immersion restraint stress. Amb Express 2017, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Lin, Q.; Huang, X.; Jiang, G.; Li, C.; Zhang, X.; Liu, S.; He, L.; Liu, Y.; Dai, Q.; et al. Effects of dietary ferulic acid on the intestinal microbiota and the associated changes on the growth performance, serum cytokine profile, and intestinal morphology in ducks. Front. Microbiol. 2021, 12, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Todokoro, T.; Negoro, H.; Kotaka, A.; Hata, Y.; Ishida, H. Aspergillus oryzae FaeA is responsible for the release of ferulic acid, a precursor of off-odor 4-vinylguaiacol in sake brewing. J. Biosci. Bioeng. 2022, 133, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Madsen, K.; Cornish, A.; Soper, P.; McKaigney, C.; Jijon, H.; Yachimec, C.; Doyle, J.; Jewell, L.; De Simone, C. Probiotic bacteria enhance murine and human intestinal epithelial barrier function. Gastroenterology 2001, 121, 580–591. [Google Scholar] [CrossRef] [Green Version]
- Leclercq, S.; Matamoros, S.; Cani, P.D.; Neyrinck, A.M.; Jamar, F.; Stärkel, P.; Windey, K.; Tremaroli, V.; Bäckhed, F.; Verbeke, K.; et al. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc. Natl. Acad. Sci. USA 2014, 111, E4485–E4493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shang, Q.; Shan, X.; Cai, C.; Hao, J.; Li, G.; Yu, G. Dietary fucoidan modulates the gut microbiota in mice by increasing the abundance of Lactobacillus and Ruminococcaceae. Food Funct. 2016, 7, 3224–3232. [Google Scholar] [CrossRef] [PubMed]
- Vijay, A.; Astbury, S.; Le Roy, C.; Spector, T.D.; Valdes, A.M. The prebiotic effects of omega-3 fatty acid supplementation: A six-week randomised intervention trial. Gut Microbes 2021, 13, 1863133. [Google Scholar] [CrossRef]
- Yan, X.; Zhang, H.; Lin, A.; Su, Y. Antagonization of Ghrelin Suppresses Muscle Protein Deposition by Altering Gut Microbiota and Serum Amino Acid Composition in a Pig Model. Biology 2022, 11, 840. [Google Scholar] [CrossRef]
- Balamurugan, R.; Chittaranjan, S.P.; Chandragunasekaran, A.M.; Ramakrishna, B.S. Molecular detection of the ruminal bacterium, Butyrivibrio fibrisolvens, in feces from rural residents of southern India. Microb. Ecol. Health Dis. 2009, 21, 38–43. [Google Scholar]
- Pane, K.; Boccella, S.; Guida, F.; Franzese, M.; Maione, S.; Salvatore, M. Role of gut microbiota in neuropathy and neuropathic pain states: A systematic preclinical review. Neurobiol. Dis. 2022, 170, 1–14. [Google Scholar] [CrossRef]
- Van der Sluis, M.; De Koning, B.A.; De Bruijn, A.C.; Velcich, A.; Meijerink, J.P.; Van Goudoever, J.B.; Büller, H.A.; Dekker, J.; Van Seuningen, I.; Renes, I.B.; et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology 2006, 131, 117–129. [Google Scholar] [CrossRef]
- Rocco, A.; Compare, D.; Angrisani, D.; Zamparelli, M.S.; Nardone, G. Alcoholic disease: Liver and beyond. World J. Gastroenterol. WJG 2014, 20, 14652. [Google Scholar] [CrossRef]
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2016, 14, 20–32. [Google Scholar] [CrossRef] [Green Version]
- Van de Wiel, A. The effect of alcohol on postprandial and fasting triglycerides. Int. J. Vasc. Med. 2012, 2012, 1–4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, S.; Xiangsong, C.; Xiang, X. Improvement of the aroma of lily rice wine by using aroma-producing yeast strain Wickerhamomyces anomalus HN006. AMB Express 2019, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ahola, A.J.; Lassenius, M.I.; Forsblom, C.; Harjutsalo, V.; Lehto, M.; Groop, P.H. Dietary patterns reflecting healthy food choices are associated with lower serum LPS activity. Sci. Rep. 2017, 7, 6511. [Google Scholar] [CrossRef] [Green Version]
- Caslin, B.; Maguire, C.; Karmakar, A.; Mohler, K.; Wylie, D.; Melamed, E. Alcohol shifts gut microbial networks and ameliorates a murine model of neuroinflammation in a sex-specific pattern. Proc. Natl. Acad. Sci. USA 2019, 116, 25808–25815. [Google Scholar] [CrossRef]
- Le Roy, C.I.; Wells, P.M.; Si, J.; Raes, J.; Bell, J.T.; Spector, T.D. Red wine consumption associated with increased gut microbiota α-diversity in 3 independent cohorts. Gastroenterology 2020, 158, 270–272. [Google Scholar] [CrossRef] [PubMed]






| Genus | Inference | p Values |
|---|---|---|
| Odoribacter | ↑ | 0.001 |
| Christensenellaceae R-7 | ↓ | 0.001 |
| Coprococcus | ↑ | 0.002 |
| Defluviitaleaceae UCG-011 | ↑ | 0.004 |
| Oscillospiraceae UCG-005 | ↑ | 0.004 |
| Rikenellaceae RC9 | ↑ | 0.006 |
| Succinivibrio | ↑ | 0.007 |
| Aeromonas | ↓ | 0.007 |
| [Eubacterium] siraeum | ↑ | 0.01 |
| Oscillospiraceae UCG-002 | ↑ | 0.013 |
| Ruminococcus | ↑ | 0.016 |
| Lachnospiraceae NK4A136 | ↑ | 0.017 |
| Lachnospiraceae NC2004 | ↑ | 0.042 |
| Butyrivibrio | ↑ | 0.023 |
| Butyricimonas | ↑ | 0.027 |
| Lachnoclostridium | ↓ | 0.03 |
| Bilophila | ↑ | 0.038 |
| Rothia | ↓ | 0.04 |
| Senegalimassilia | ↑ | 0.045 |
| Anaerovoracaceae Family XIII UCG-001 | ↑ | 0.047 |
| Treatment | Bacteria | Inference | p Values |
|---|---|---|---|
| CT | Eubacterium ventriosum | ↑ | 0.037 |
| Lachnospiraceae A2 | ↓ | 0.037 | |
| Cuneatibacter | ↓ | 0.037 | |
| Leptotrichia | ↓ | 0.046 | |
| Negativibacillus | ↓ | 0.046 | |
| Mesocricetibacter | ↓ | 0.046 | |
| Rodentibacter | ↓ | 0.046 | |
| Ruminococcus | ↑ | 0.046 | |
| Candidatus Arthromitus | ↓ | 0.050 | |
| Intestinimonas | ↓ | 0.050 | |
| Lachnospiraceae NK4A136 | ↑ | 0.050 | |
| Odoribacter | ↓ | 0.050 | |
| RB | Eubacterium siraeum | ↑ | 0.037 |
| Anaerovoracaceae Family XIII UCG-001 | ↓ | 0.037 | |
| Ruminococcaceae UBA1819 | ↑ | 0.037 | |
| Eubacterium nodatum | ↓ | 0.046 | |
| Lachnospiraceae ASF356 | ↓ | 0.046 | |
| Rodentibacter | ↓ | 0.046 | |
| Anaerovoracaceae Family XIII AD3011 | ↓ | 0.046 | |
| Streptococcus | ↓ | 0.046 | |
| Negativibacillus | ↓ | 0.046 | |
| Roseburia | ↓ | 0.050 | |
| Acetatifactor | ↑ | 0.050 | |
| Bacteroides | ↓ | 0.050 | |
| Ligilactobacillus | ↓ | 0.050 | |
| Anaerotruncus | ↓ | 0.050 | |
| Erysipelatoclostridium | ↓ | 0.050 | |
| Prevotellaceae UCG-001 | ↑ | 0.050 | |
| IF | Lachnospiraceae ASF356 | ↑ | 0.000 |
| Paludicola | ↑ | 0.000 | |
| Anaerovoracaceae Family XIII AD3011 | ↓ | 0.001 | |
| Parasporobacterium | ↓ | 0.001 | |
| SF | Wautersiella | ↑ | 0.037 |
| Leptotrichia | ↓ | 0.046 | |
| Candidatus Stoquefichus | ↓ | 0.046 | |
| Lachnospiraceae UCG-004 | ↓ | 0.050 | |
| Lachnospiraceae ASF356 | ↑ | 0.050 | |
| Rikenella | ↑ | 0.050 | |
| Desulfovibrio | ↑ | 0.050 | |
| Lactobacillaceae HT002 | ↓ | 0.050 | |
| Intestinimonas | ↑ | 0.050 | |
| Bacteroides | ↓ | 0.050 | |
| Lactobacillus | ↓ | 0.050 | |
| Ligilactobacillus | ↑ | 0.050 | |
| Eubacterium xylanophilum | ↓ | 0.050 | |
| Prevotellaceae UCG-001 | ↑ | 0.050 | |
| Ruminococcus | ↑ | 0.050 |
| Metabolites | CT | RB | IF | SF |
|---|---|---|---|---|
| alpha Linolenic acid | 2.54 | 0.955 | 0.505 | 0.13 |
| 17-Octadecynoic acid | 0 | 0.075 | 0 | 0 |
| 1-Monolinoleoylglycerol | 0.75 | 2.59 | 0.205 | 3.075 |
| 1-O-Heptadecylglycerol | 0 | 0 | 0 | 0.03 |
| 1-O-hexadecylglycerol | 0 | 0.2 | 0.255 | 0.595 |
| 2-alpha -Mannobiose | 0 | 0.13 | 0 | 0 |
| 2-Desoxy-pentos-3-ulose | 0.045 | 0.11 | 0.095 | 0.175 |
| 2-Pyrrolidone-5-carboxylic acid | 0.525 | 0.07 | 0.43 | 0.455 |
| 3-Hydroxydodecanedioic acid | 0.17 | 0 | 0 | 0 |
| 4-Hydroxyphenylbutyric acid | 0 | 0.155 | 0.235 | 0 |
| 4-Nitrophenyl-.beta.-D-galacturonide | 0 | 0.065 | 0 | 0 |
| 5-Methyluridine | 0 | 0 | 0.275 | 0 |
| 9,12,15-Octadecatrienoic acid | 0 | 0 | 0.1 | 0 |
| 9-Octadecenoic acid | 0 | 2.1 | 2.645 | 1.485 |
| Acetic acid | 0.14 | 0.44 | 0.11 | 0.215 |
| alpha-L-Mannopyranose | 0 | 0.025 | 0 | 0 |
| Arachidonic acid | 0 | 0 | 0 | 0.32 |
| Benzenepropanoic acid | 0.16 | 0 | 0.11 | 0 |
| Benzoic acid | 0 | 0 | 0 | 0.17 |
| Butanedioic acid | 1.91 | 0.725 | 1.07 | 3.045 |
| Butanoic acid | 0 | 0 | 1.225 | 0.11 |
| Cinnamate | 0 | 0.025 | 0 | 0.03 |
| D-(-)-Erythrose | 0 | 0.015 | 0 | 0 |
| D-(-)-Rhamnose | 0 | 1.04 | 1.905 | 2.625 |
| D-(-)-Tagatose | 0 | 0.11 | 0.345 | 0.195 |
| D-(+)-Cellobiose | 0 | 0.885 | 0.76 | 0.595 |
| D-(+)-Glucuronic acid | 0 | 0.07 | 0.075 | 0 |
| D-(+)-Xylose | 8.415 | 5.38 | 9.36 | 5.79 |
| D-Arabinonic acid | 0 | 0 | 0 | 0.02 |
| D-Galactose | 16.96 | 12.04 | 11.11 | 13.67 |
| D-Glucitol | 0 | 0 | 0 | 1.315 |
| D-Glucose | 0 | 16.4 | 0 | 0 |
| D-Lactose | 0 | 2.015 | 0 | 0 |
| D-Mannose | 0.34 | 0.215 | 0 | 0.895 |
| Dodecanedioic acid | 0 | 0.12 | 0.285 | 0 |
| D-Xylose | 0 | 0 | 0 | 0.05 |
| Eicosanoic acid | 0 | 0 | 0 | 0.095 |
| Erythro-Pentonic acid | 0 | 0.04 | 0.21 | 0.05 |
| Ethanedioic acid | 0.4 | 0 | 0.955 | 0.845 |
| Ethanol | 0 | 0 | 0 | 0.01 |
| Glycerol | 4.41 | 4.84 | 4.31 | 5.295 |
| Glycine | 0 | 0 | 0 | 0.035 |
| Hexadecanoic acid | 0 | 7.495 | 6.225 | 5.85 |
| L-(-)-Sorbose | 0 | 0 | 0 | 0.02 |
| L-Asparagine | 0 | 0 | 0 | 0.155 |
| L-Isoleucine | 0.07 | 0.03 | 0.125 | 0.35 |
| L-Threonine | 0 | 0 | 0.25 | 0.79 |
| L-Valine | 0.135 | 0.1 | 0.2 | 0.355 |
| Maltose | 0 | 0.155 | 0 | 0 |
| Monoamidoethylmalonic acid | 0 | 0 | 0.01 | 0 |
| N-Acetyl-D-galactosaminitol | 0 | 0 | 0 | 1.07 |
| N-Acetyl-D-glucosamine | 2.955 | 1.415 | 4.525 | 4.78 |
| n-Pentadecanoic acid | 0 | 0 | 0 | 0.27 |
| N-phthalimide | 1.415 | 1.355 | 0 | 2.44 |
| Octadecanoic acid | 0 | 4.4 | 0 | 0 |
| Oleic acid | 7.62 | 5.125 | 4.19 | 4.84 |
| Pentanedioic acid | 0.045 | 0 | 0.015 | 0.015 |
| Pentanoic acid | 0 | 0 | 0 | 1.22 |
| Phenyl alanine | 0 | 0 | 0 | 0.045 |
| Phosphoric acid | 0 | 0 | 0.04 | 0 |
| Propanedioic acid | 0 | 0 | 0 | 0.12 |
| Propanetriol | 0 | 0.015 | 0.03 | 0.04 |
| Propanoic acid | 9.89 | 7.06 | 6.075 | 13.01 |
| Ribitol | 2.38 | 0.03 | 0 | 0 |
| Serine | 0.1 | 0.04 | 0.19 | 0.265 |
| Tetracosanoic acid | 0 | 0.18 | 0.28 | 0.42 |
| Tetradecanoic acid | 0.065 | 0 | 0.395 | 0.2 |
| Uridine | 0 | 0 | 0.105 | 0.04 |
| CT | RB | IF | SF | ||
|---|---|---|---|---|---|
| Villus length (m) | 169.852 ± 5.92 | 151.848 ± 3.59 | 155.89 ± 10.73 | 176.914 ± 9.92 | |
| Crypt depth (m) | 84.022 ± 1.96 | 82.892 ± 2.98 | 74.65 ± 2.21 | 93.246 ± 2.39 | |
| Width of lamina propria (m) | 26.94 ± 2.64 | 27.236 ± 1.80 | 30.108 ± 2.86 | 31.902 ± 1.33 | |
| Depth of submucosa (m) | 22.51 ± 1.44 | 25.376 ± 3.00 | 21.23 ± 1.64 | 29.43 ± 2.35 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhaskar, B.; Bhattacharya, A.; Adak, A.; Das, S.; Khan, M.R. A Human and Animal Based Study Reveals That a Traditionally Fermented Rice Beverage Alters Gut Microbiota and Fecal Metabolites for Better Gut Health. Fermentation 2023, 9, 126. https://doi.org/10.3390/fermentation9020126
Bhaskar B, Bhattacharya A, Adak A, Das S, Khan MR. A Human and Animal Based Study Reveals That a Traditionally Fermented Rice Beverage Alters Gut Microbiota and Fecal Metabolites for Better Gut Health. Fermentation. 2023; 9(2):126. https://doi.org/10.3390/fermentation9020126
Chicago/Turabian StyleBhaskar, Bhuwan, Anupam Bhattacharya, Atanu Adak, Santanu Das, and Mojibur R. Khan. 2023. "A Human and Animal Based Study Reveals That a Traditionally Fermented Rice Beverage Alters Gut Microbiota and Fecal Metabolites for Better Gut Health" Fermentation 9, no. 2: 126. https://doi.org/10.3390/fermentation9020126
APA StyleBhaskar, B., Bhattacharya, A., Adak, A., Das, S., & Khan, M. R. (2023). A Human and Animal Based Study Reveals That a Traditionally Fermented Rice Beverage Alters Gut Microbiota and Fecal Metabolites for Better Gut Health. Fermentation, 9(2), 126. https://doi.org/10.3390/fermentation9020126