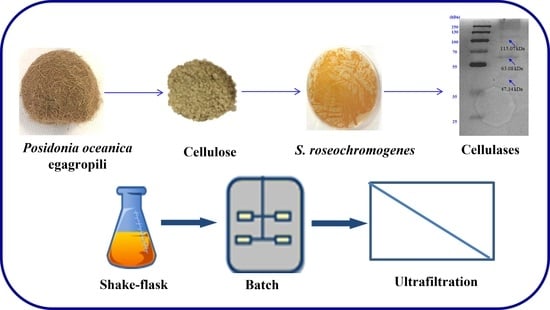
Cellulose from Posidonia oceanica Sea Balls (Egagropili) as Substrate to Enhance Streptomyces roseochromogenes Cellulase Biosynthesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Microorganism and Medium
2.3. Shake Flask Experiments
2.4. Batch Experiments
2.5. Extracellular Cellulase Recovery by Ultrafiltration
2.6. Protein Content Determination
2.7. Monosaccharide Determination
2.8. SDS-PAGE and Zymogram Analyses
2.9. Determination of the Optimal Conditions of Cellulase Activity
2.10. Cellulase Activity Assays
3. Results
3.1. Shake Flask Experiments and Cellulase Purification by Ultrafiltration Membranes
3.2. Determination of Cellulase Molecular Weights and Optimal Activity Conditions
3.3. Cellulase Activity Assays
3.4. Batch Experiments
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barbuto Ferraiuolo, S.; Cammarota, M.; Schiraldi, C.; Restaino, O.F. Streptomycetes as platform for biotechnological production processes of drugs. Appl. Microbiol. Biotechnol. 2021, 105, 551–568. [Google Scholar] [CrossRef] [PubMed]
- Leo, V.V.; Asem, D.; Zothanpuia Singh, B.P. Actinobacteria: A highly potent source for holocellulose degrading enzymes. In New and Future Development in Microbial Biotechnology and Bioengineering-Actinobacteria: Diversity and Biotechnological Applications; Singh, B.P., Gupta, V.K., Passar, A.K., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 191–205. [Google Scholar]
- Kuhad, R.C.; Gupta, R.; Singh, A. Microbial cellulases and their industrial applications. Enzym. Res. 2011, 2011, 280696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ventorino, V.; Ionata, E.; Birolo, L.; Montella, S.; Marcolongo, L.; De Chiaro, A.; Espresso, F.; Faraco, V.; Pepe, O. Lignocellulose-adapted endo-cellulase producing Streptomyces strains for bioconversion of cellulose-based materials. Front. Microbiol. 2016, 7, 2061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Vinha, F.N.M.; Gravina-Oliveira, M.P.; Franco, M.N.; Macrae, A.; da Silva Bon, E.P.; Nascimento, R.P.; Coelho, R.R.R. Cellulase production by Streptomyces viridobrunneus SCPE-09 using lignocellulosic biomass as inducer substrate. Appl. Biochem. Biotechnol. 2011, 164, 256–267. [Google Scholar] [CrossRef] [PubMed]
- Celaya-Herrera, S.; Casados-Vázquez, L.E.; Valdez-Vazquez, I.; Barona-Gómez, F.; Bideshi, D.K.; Barboza-Corona, J.E. A cellulolytic Streptomyces sp. isolated from a highly oligotrophic niche shows potential for hydrolyzing agricultural wastes. BioEnergy Res. 2021, 14, 333–343. [Google Scholar] [CrossRef]
- Lim, J.H.; Lee, C.R.; Dhakshnamoorthy, V.; Park, J.S.; Hong, S.K. Molecular characterization of Streptomyces coelicolor A (3) SCO6548 as a cellulose 1,4-β-cellobiosidase. FEMS Microbiol. Lett. 2016, 363, fnv245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Sersy, N.A.; Abd-Elnaby, H.; Abou-Elela, G.M.; Ibrahim, H.A.; El-Toukhy, N.M. Optimization, economization and characterization of cellulase produced by marine Streptomyces ruber. Afr. J. Biotechnol. 2010, 9, 6355–6364. [Google Scholar]
- Prasad, P.; Singh, T.; Bedi, S. Characterization of the cellulolytic enzyme produced by Streptomyces griseorubens isolated from Indian soil. J. King Saud Univ.-Sci. 2013, 25, 245–250. [Google Scholar] [CrossRef] [Green Version]
- Restaino, O.F.; Marseglia, M.; De Castro, C.; Diana, P.; Forni, P.; Parrilli, M.; De Rosa, M.; Schiraldi, C. Biotechnological transformation of hydrocortisone to 16α-hydroxy hydrocortisone by Streptomyces roseochromogenes. Appl. Microbiol. Biotechnol. 2014, 98, 1291–1299. [Google Scholar] [CrossRef]
- Restaino, O.F.; Marseglia, M.; Diana, P.; Borzacchiello, M.G.; Finamore, R.; Vitiello, M.; D’Agostino, A.; De Rosa, M.; Schiraldi, C. Advances in the 16α-hydroxy transformation of hydrocortisone by Streptomyces roseochromogenes. Process. Biochem. 2016, 51, 1–8. [Google Scholar] [CrossRef]
- Restaino, O.F.; Barbuto Ferraiuolo, S.; Perna, A.; Cammarota, M.; Borzacchiello, M.G.; Fiorentino, A.; Schiraldi, C. Biotechnological transformation of hydrocortisone into 16α-hydroxyprednisolone by coupling Arthrobacter simplex and Streptomyces roseochromogenes. Molecules 2020, 25, 4912. [Google Scholar] [CrossRef] [PubMed]
- Restaino, O.F.; Scognamiglio, M.; Mirpoor, S.F.; Cammarota, M.; Ventriglia, R.; Giosafatto, C.V.L.; Fiorentino, A.; Porta, R.; Schiraldi, C. Enhanced Streptomyces roseochromogenes melanin production by using the marine renewable source Posidonia oceanica egagropili. Appl. Microbiol. Biotechnol. 2022, 106, 7265–7283. [Google Scholar] [CrossRef] [PubMed]
- Mirpoor, S.F.; Giosafatto, C.V.L.; Di Pierro, P.; Di Girolamo, R.; Regalado-González, C.; Porta, R. Valorisation of Posidonia oceanica sea balls (egagropili) as a potential source of reinforcement agents in protein-based biocomposites. Polymers 2020, 12, 2788. [Google Scholar] [CrossRef] [PubMed]
- Mirpoor, S.F.; Restaino, O.F.; Schiraldi, C.; Giosafatto, C.V.L.; Ruffo, F.; Porta, R. Lignin/carbohydrate complex isolated from Posidonia oceanica sea balls (Egagropili): Characterization and antioxidant reinforcement of protein-based films. Int. J. Mol. Sci. 2021, 22, 9147. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Restaino, O.F.; Borzacchiello, M.G.; Scognamiglio, I.; Porzio, E.; Manco, G.; Fedele, L.; Donatiello, C.; De Rosa, M.; Schiraldi, C. Boosted large-scale production and purification of a thermostable archeal phosphotriesterase-like lactonase for organophosphate decontamination. J. Ind. Microbiol. Biotechnol. 2017, 44, 363–375. [Google Scholar] [CrossRef]
- Restaino, O.F.; Borzacchiello, M.G.; Scognamiglio, I.; Fedele, L.; Alfano, A.; Porzio, E.; Manco, G.; Donatiello, C.; De Rosa, M.; Schiraldi, C. High yield production and purificaton of two recombinant thermostable phosphotriesterase-like lactonases from Sulfolobus acidocaldarius and Sulfolobus solfataricus useful as bioremediation tools and bioscavengers. BMC Biotechnol. 2018, 18, 18. [Google Scholar] [CrossRef] [Green Version]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Hsu, C.L.; Chang, K.S.; Lai, M.Z.; Chang, T.C.; Chang, Y.H.; Jang, H.D. Pretreatment and hydrolysis of cellulosic agricultural wastes with a cellulase-producing Streptomyces for bioethanol production. Biomass Bioenergy 2011, 35, 1878–1884. [Google Scholar] [CrossRef]
- Berlin, A.; Gilkes, N.; Kilburn, D.; Maximenko, V.; Bura, R.; Markov, A.; Skomarovsky, A.; Gusakov, A.; Sinitsyn, A.; Okunev, O.; et al. Evaluation of cellulase preparations for hydrolysis of hardwood substrates. Appl. Biochem. Biotechnol. 2006, 130, 528–545. [Google Scholar] [CrossRef]
- Merino, S.T.; Cherry, J. Progress and challenges in enzyme development for biomass utilization. In Advances in Biochemical Engineering/Biotechnology; Olsson, L., Ed.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 108. [Google Scholar] [CrossRef]
- Takasuka, T.E.; Book, A.J.; Lewin, G.R.; Currie, C.R.; Fox, B.G. Aerobic deconstruction of cellulosic biomass by an insect-associated Streptomyces. Sci. Rep. 2013, 3, 1030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segato, F.; Dama’sio, A.R.L.; de Lucas, R.C.; Squina, F.M.; Prade, R.A. Genomics review of holocellulose deconstruction by Aspergilli. Microbiol. Mol. Biol. Rev. 2014, 78, 588–613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bispo, A.S.R.; Andrade, J.P.; Souza, D.T.; Teles, Z.N.S.; Nascimento, R.P. Utilization of agroindustrial by-products as substrate in endoglucanase production by Streptomyces diastaticus PA-01 under submerged fermentation. Braz. J. Chem. Eng. 2018, 35, 429–440. [Google Scholar] [CrossRef] [Green Version]
- de Lima, A.L.G.; do Nascimento, R.P.; da Silva Bon, E.P.; Coelho, R.R.R. Streptomyces drozdowiczii cellulase production using agro-industrial by-products and its potential use in the detergent and textile industries. Enzym. Microb. Technol. 2005, 37, 272–277. [Google Scholar] [CrossRef]






| Sample | Volume (L) | Protein (g·L−1) | Activity (U·L−1) | Specific Activity (U·g−1) | Purification Fold |
|---|---|---|---|---|---|
| Supernatant of control growth w/o cellulose | 0.400 | 0.227 | 103 | 458 | |
| 10 kDa retentate of control growth w/o cellulose | 0.033 | 0.296 | 3429 | 11584 | 25 |
| Supernatant of growth with cellulose | 0.400 | 0.242 | 267 | 1103 | |
| 10 kDa retentate of growth with cellulose | 0.026 | 0.348 | 8573 | 24635 | 22 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Restaino, O.F.; Cuomo, S.; D’Ambrosio, S.; Vassallo, V.; Mirpoor, S.F.; Giosafatto, C.V.L.; Porta, R.; Schiraldi, C. Cellulose from Posidonia oceanica Sea Balls (Egagropili) as Substrate to Enhance Streptomyces roseochromogenes Cellulase Biosynthesis. Fermentation 2023, 9, 98. https://doi.org/10.3390/fermentation9020098
Restaino OF, Cuomo S, D’Ambrosio S, Vassallo V, Mirpoor SF, Giosafatto CVL, Porta R, Schiraldi C. Cellulose from Posidonia oceanica Sea Balls (Egagropili) as Substrate to Enhance Streptomyces roseochromogenes Cellulase Biosynthesis. Fermentation. 2023; 9(2):98. https://doi.org/10.3390/fermentation9020098
Chicago/Turabian StyleRestaino, Odile Francesca, Sabrina Cuomo, Sergio D’Ambrosio, Valentina Vassallo, Seyedeh Fatemeh Mirpoor, Concetta Valeria L. Giosafatto, Raffaele Porta, and Chiara Schiraldi. 2023. "Cellulose from Posidonia oceanica Sea Balls (Egagropili) as Substrate to Enhance Streptomyces roseochromogenes Cellulase Biosynthesis" Fermentation 9, no. 2: 98. https://doi.org/10.3390/fermentation9020098
APA StyleRestaino, O. F., Cuomo, S., D’Ambrosio, S., Vassallo, V., Mirpoor, S. F., Giosafatto, C. V. L., Porta, R., & Schiraldi, C. (2023). Cellulose from Posidonia oceanica Sea Balls (Egagropili) as Substrate to Enhance Streptomyces roseochromogenes Cellulase Biosynthesis. Fermentation, 9(2), 98. https://doi.org/10.3390/fermentation9020098