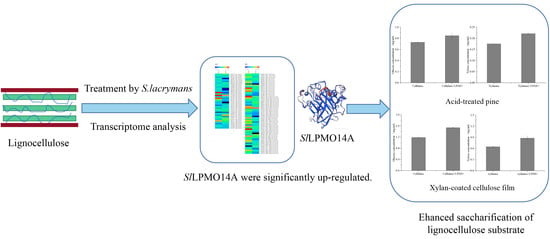
Lytic Polysaccharide Monooxygenases from Serpula lacrymans as Enzyme Cocktail Additive for Efficient Lignocellulose Degradation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Medium
2.2. Transcriptome Analysis of Serpula Lacrymans Incubated on Pine
2.3. Sequence Alignment and Phylogenetic Tree Analysis of SlLPMO14A
2.4. Expression and Purification of Recombinant SlLPMO14A
2.5. Enzymatic Properties of SlLPMO14A
2.6. Polysaccharides Cleavage, ESEM and AFM Analysis
2.7. Saccharification Assays
3. Results and Discussion
3.1. Transcriptomic Analysis Reveals Co-Regulation of SlLPMO14A with the Genes Involved in Fenton Reaction and Polysaccharides Degradation in S. lacrymans
3.2. The Expression of SlLPMO14A and Enzymatic Properties of SlLPMO14A
3.3. Polysaccharides Cleavage and the Synergy Assays for the Saccharification of Biomass
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Quinlan, R.J.; Sweeney, M.D.; Lo Leggio, L.; Otten, H.; Poulsen, J.C.N.; Johansen, K.S.; Krogh, K.; Jorgensen, C.I.; Tovborg, M.; Anthonsen, A.; et al. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc. Natl. Acad. Sci. USA 2011, 108, 15079–15084. [Google Scholar] [CrossRef] [PubMed]
- Harris, P.V.; Welner, D.; McFarland, K.C.; Re, E.; Poulsen, J.C.N.; Brown, K.; Salbo, R.; Ding, H.S.; Vlasenko, E.; Merino, S.; et al. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: Structure and function of a large, enigmatic family. Biochemistry 2010, 49, 3305–3316. [Google Scholar] [CrossRef] [PubMed]
- Kubicek, C.P.; Kubicek, E.M. Enzymatic deconstruction of plant biomass by fungal enzymes. Curr. Opin. Chem. Biol. 2016, 35, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Vandhana, T.M.; Reyre, J.L.; Sushmaa, D.; Berrin, J.G.; Bissaro, B.; Madhuprakash, J. On the expansion of biological functions of lytic polysaccharide monooxygenases. New Phytol. 2022, 233, 2380–2396. [Google Scholar] [CrossRef]
- Tandrup, T.; Frandsen, K.E.H.; Johansen, K.S.; Berrin, J.G.; Lo Leggio, L. Recent insights into lytic polysaccharide monooxygenases (LPMOs). Biochem. Soc. Trans. 2018, 46, 1431–1447. [Google Scholar] [CrossRef]
- Zhang, J.W.; Presley, G.N.; Hammel, K.E.; Ryu, J.S.; Menke, J.R.; Figueroa, M.; Hu, D.H.; Orr, G.; Schilling, J.S. Localizing gene regulation reveals a staggered wood decay mechanism for the brown rot fungus Postia placenta . Proc. Natl. Acad. Sci. USA 2016, 113, 10968–10973. [Google Scholar] [CrossRef]
- Goodell, B.; Jellison, J.; Liu, J.; Daniel, G.; Paszczynski, A.; Fekete, F.; Krishnamurthy, S.; Jun, L.; Xu, G. Low molecular weight chelators and phenolic compounds isolated from wood decay fungi and their role in the fungal biodegradation of wood. J. Biotechnol. 1997, 53, 133–162. [Google Scholar] [CrossRef]
- Ray, M.J.; Leak, D.J.; Spanu, P.D.; Murphy, R.J. Brown rot fungal early stage decay mechanism as a biological pretreatment for softwood biomass in biofuel production. Biomass Bioenergy 2010, 34, 1257–1262. [Google Scholar] [CrossRef]
- Martinez, D.; Challacombe, J.; Morgenstern, I.; Hibbett, D.; Schmoll, M.; Kubicek, C.P.; Ferreira, P.; Ruiz-Duenas, F.J.; Martinez, A.T.; Kersten, P.; et al. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc. Natl. Acad. Sci. USA 2009, 106, 1954–1959. [Google Scholar] [CrossRef]
- Hori, C.; Gaskell, J.; Igarashi, K.; Samejima, M.; Hibbett, D.; Henrissat, B.; Cullen, D. Genomewide analysis of polysaccharides degrading enzymes in 11 white- and brown-rot Polyporales provides insight into mechanisms of wood decay. Mycologia 2013, 105, 1412–1427. [Google Scholar] [CrossRef]
- Floudas, D.; Binder, M.; Riley, R.; Barry, K.; Blanchette, R.A.; Henrissat, B.; Martinez, A.T.; Otillar, R.; Spatafora, J.W.; Yadav, J.S.; et al. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 2012, 336, 1715–1719. [Google Scholar] [CrossRef] [PubMed]
- Eastwood, D.C. The plant cell wall-decomposing machinery underlies the functional diversity of forest fungi. Science 2011, 333, 762–765. [Google Scholar] [CrossRef] [PubMed]
- Kojima, Y.; Varnai, A.; Ishida, T.; Sunagawa, N.; Petrovic, D.M.; Igarashi, K.; Jellison, J.; Goodell, B.; Alfredsen, G.; Westereng, B.; et al. A lytic polysaccharide monooxygenase with broad xyloglucan specificity from the brown-rot fungus Gloeophyllum trabeum and its action on cellulose-xyloglucan complexes. Appl. Environ. Microbiol. 2016, 82, 6557–6572. [Google Scholar] [CrossRef] [PubMed]
- Couturier, M.; Ladeveze, S.; Sulzenbacher, G.; Ciano, L.; Fanuel, M.; Moreau, C.; Villares, A.; Cathala, B.; Chaspoul, F.; Frandsen, K.E.; et al. Lytic xylan oxidases from wood-decay fungi unlock biomass degradation. Nat. Chem. Biol. 2018, 14, 306–310. [Google Scholar] [CrossRef]
- Zerva, A.; Pentari, C.; Grisel, S.; Berrin, J.G.; Topakas, E. A new synergistic relationship between xylan-active LPMO and xylobiohydrolase to tackle recalcitrant xylan. Biotechnol. Biofuels 2020, 13, 142. [Google Scholar] [CrossRef]
- Mahajan, R.; Hudson, B.S.; Sharma, D.; Kolte, V.; Sharma, G.; Goel, G. Transcriptome analysis of Podoscypha petalodes strain GGF6 reveals the diversity of proteins involved in lignocellulose degradation and ligninolytic function. Indian J. Microbiol. 2022, 62, 569–582. [Google Scholar] [CrossRef]
- Forsberg, Z.; Vaaje-Kolstad, G.; Westereng, B.; Bunaes, A.C.; Stenstrom, Y.; MacKenzie, A.; Sorlie, M.; Horn, S.J.; Eijsink, V.G.H. Cleavage of cellulose by a CBM33 protein. Protein Sci. 2011, 20, 1479–1483. [Google Scholar] [CrossRef]
- Li, F.; Zhao, H.L.; Liu, Y.X.; Zhang, J.Q.; Yu, H.B. Chitin biodegradation by lytic polysaccharide monooxygenases from Streptomyces coelicolor in vitro and in vivo. Int. J. Mol. Sci. 2023, 24, 275. [Google Scholar] [CrossRef]
- Loose, J.S.M.; Forsberg, Z.; Fraaije, M.W.; Eijsink, V.G.H.; Vaaje-Kolstad, G. A rapid quantitative activity assay shows that the Vibrio cholerae colonization factor GbpA is an active lytic polysaccharide monooxygenase. FEBS Lett. 2014, 588, 3435–3440. [Google Scholar] [CrossRef]
- Kittl, R.; Kracher, D.; Burgstaller, D.; Haltrich, D.; Ludwig, R. Production of four Neurospora crassa lytic polysaccharide monooxygenases in Pichia pastoris monitored by a fluorimetric assay. Biotechnol. Biofuels 2012, 5, 79. [Google Scholar] [CrossRef]
- Breslmayr, E.; Hanzek, M.; Hanrahan, A.; Leitner, C.; Kittl, R.; Santek, B.; Oostenbrink, C.; Ludwig, R. A fast and sensitive activity assay for lytic polysaccharide monooxygenase. Biotechnol. Biofuels 2018, 11, 79. [Google Scholar] [CrossRef]
- Siqueira, G.; Varnai, A.; Ferraz, A.; Milagres, A.M.F. Enhancement of cellulose hydrolysis in sugarcane bagasse by the selective removal of lignin with sodium chlorite. Appl. Energy 2013, 102, 399–402. [Google Scholar] [CrossRef]
- Ni, H.X.; Li, M.J.; Li, F.; Wang, L.; Xie, S.X.; Zhang, X.Y.; Yu, H.B. In-situ lignin drives lytic polysaccharide monooxygenases to enhance enzymatic saccharification. Int. J. Biol. Macromol. 2020, 161, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Ma, F.Y.; Zhao, H.L.; Zhang, S.; Wang, L.; Zhang, X.Y.; Yu, H.B. A lytic polysaccharide monooxygenase from a white-rot fungus drives the degradation of lignin by a versatile peroxidase. Appl. Environ. Microbiol. 2019, 85, e02803–e02818. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Sun, X.J.; Yu, W.; Shi, C.C.; Zhang, X.Y.; Yu, H.B.; Ma, F. Enhanced konjac glucomannan hydrolysis by lytic polysaccharide monooxygenases and generating prebiotic oligosaccharides. Carbohydr. Polym. 2021, 253, 117241. [Google Scholar] [CrossRef] [PubMed]
- Brenelli, L.; Squina, F.M.; Felby, C.; Cannella, D. Laccase-derived lignin compounds boost cellulose oxidative enzymes AA9. Biotechnol. Biofuels 2018, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Plaza, N.; Kojima, Y.; Yoshida, M.; Zhang, J.W.; Jellison, J.; Pingali, S.V.; O’Neill, H.; Goodell, B. Nanostructural analysis of enzymatic and non-enzymatic brown rot fungal deconstruction of the lignocellulose cell wall. Front. Microbiol. 2020, 11, 1389. [Google Scholar] [CrossRef] [PubMed]
- Manavalan, T.; Stepnov, A.A.; Hegnar, O.A.; Eijsink, V.G.H. Sugar oxidoreductases and LPMOs-two sides of the same polysaccharide degradation story? Carbohydr. Res. 2021, 505, 108350. [Google Scholar] [CrossRef]
- Kracher, D.; Scheiblbrandner, S.; Felice, A.K.G.; Breslmayr, E.; Preims, M.; Ludwicka, K.; Haltrich, D.; Eijsink, V.G.H.; Ludwig, R. Extracellular electron transfer systems fuel cellulose oxidative degradation. Science 2016, 352, 1098–1101. [Google Scholar] [CrossRef]
- Bissaro, B.; Rohr, A.K.; Muller, G.; Chylenski, P.; Skaugen, M.; Forsberg, Z.; Horn, S.J.; Vaaje-Kolstad, G.; Eijsink, V.G.H. Oxidative cleavage of polysaccharides by monocopper enzymes depends on H2O2. Nat. Chem. Biol. 2017, 13, 1123–1128. [Google Scholar] [CrossRef]
- Serdakowski, A.L.; Dordick, J.S. Enzyme activation for organic solvents made easy. Trends Biotechnol. 2008, 26, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Gangadhara; Kumar, P.; Prakash, V. Influence of polyols on the stability and kinetic parameters of invertase from Candida utilis: Correlation with the conformational stability and activity. Protein J. 2008, 27, 440–449. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, F.; Liu, Y.; Zhao, H.; Liu, X.; Yu, H. Lytic Polysaccharide Monooxygenases from Serpula lacrymans as Enzyme Cocktail Additive for Efficient Lignocellulose Degradation. Fermentation 2023, 9, 506. https://doi.org/10.3390/fermentation9060506
Li F, Liu Y, Zhao H, Liu X, Yu H. Lytic Polysaccharide Monooxygenases from Serpula lacrymans as Enzyme Cocktail Additive for Efficient Lignocellulose Degradation. Fermentation. 2023; 9(6):506. https://doi.org/10.3390/fermentation9060506
Chicago/Turabian StyleLi, Fei, Yang Liu, Honglu Zhao, Xuan Liu, and Hongbo Yu. 2023. "Lytic Polysaccharide Monooxygenases from Serpula lacrymans as Enzyme Cocktail Additive for Efficient Lignocellulose Degradation" Fermentation 9, no. 6: 506. https://doi.org/10.3390/fermentation9060506
APA StyleLi, F., Liu, Y., Zhao, H., Liu, X., & Yu, H. (2023). Lytic Polysaccharide Monooxygenases from Serpula lacrymans as Enzyme Cocktail Additive for Efficient Lignocellulose Degradation. Fermentation, 9(6), 506. https://doi.org/10.3390/fermentation9060506